
uncultured phage_MedDCM-OCT-S30-C28
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Fussvirus; Fussvirus S30C28
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
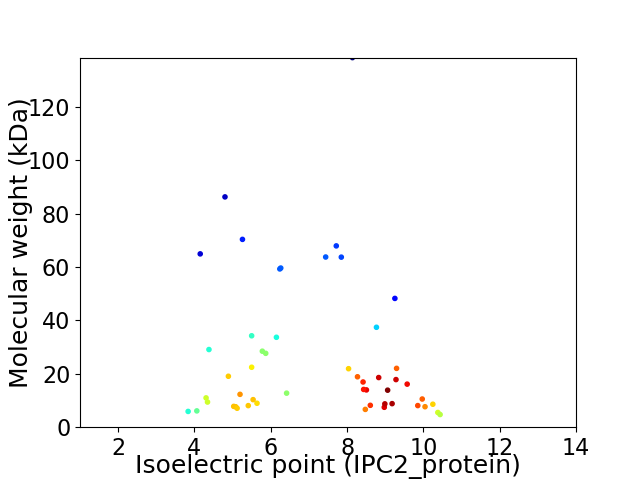
Virtual 2D-PAGE plot for 51 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6S4PDV3|A0A6S4PDV3_9CAUD Uncharacterized protein OS=uncultured phage_MedDCM-OCT-S30-C28 OX=2741076 PE=4 SV=1
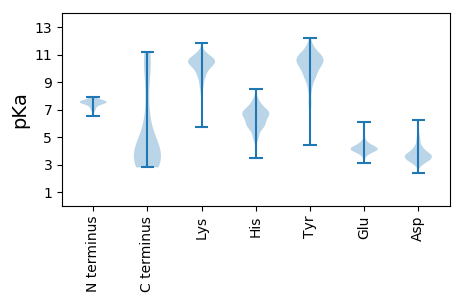
MM1 pKa = 7.74SYY3 pKa = 10.43LAQVSYY9 pKa = 10.36TGDD12 pKa = 3.32GSTTQYY18 pKa = 11.16SITFPFLDD26 pKa = 3.73STHH29 pKa = 5.29VKK31 pKa = 10.74AFIGGVATTAFTISSSTLTFTSAPANSAVIRR62 pKa = 11.84IEE64 pKa = 3.86RR65 pKa = 11.84QTPTSSRR72 pKa = 11.84LVDD75 pKa = 4.11FTDD78 pKa = 4.41GSVLTEE84 pKa = 3.91SDD86 pKa = 3.6LDD88 pKa = 3.76RR89 pKa = 11.84SADD92 pKa = 3.47QNFYY96 pKa = 10.3IAQEE100 pKa = 3.83ISDD103 pKa = 4.29ANQSAMTIGTDD114 pKa = 4.3DD115 pKa = 4.6KK116 pKa = 11.5FDD118 pKa = 3.85AQSKK122 pKa = 9.01IIKK125 pKa = 9.87NVANPVDD132 pKa = 3.74NQDD135 pKa = 3.81AVTKK139 pKa = 10.51YY140 pKa = 10.39YY141 pKa = 11.04LEE143 pKa = 4.28NTWLSSANKK152 pKa = 8.63TALTTVNSNITAITNVNSNSTNINAVNSNSTNINLVAGSIGSVNTVATDD201 pKa = 3.31IAKK204 pKa = 10.49VIAVANDD211 pKa = 3.2LAEE214 pKa = 4.27AVSEE218 pKa = 4.53VEE220 pKa = 4.57TVANDD225 pKa = 3.74LNEE228 pKa = 4.33STSEE232 pKa = 3.68IDD234 pKa = 3.35TVANAITNVDD244 pKa = 3.0IVGNAIANVNTLAGVSNLQNLANAHH269 pKa = 5.0SQIVTVGNNIPAVEE283 pKa = 4.14AFGNTYY289 pKa = 9.96KK290 pKa = 10.42ISASAPSSPIEE301 pKa = 3.8GTLWFDD307 pKa = 3.32TTADD311 pKa = 3.03IMKK314 pKa = 9.86VYY316 pKa = 10.5DD317 pKa = 3.6GSSFVNAGSSVNGTSGRR334 pKa = 11.84FKK336 pKa = 11.11YY337 pKa = 10.11IATAGQTTFTGASHH351 pKa = 7.24SDD353 pKa = 3.16TGSNTLTYY361 pKa = 10.48DD362 pKa = 2.98AGFIDD367 pKa = 4.02VFKK370 pKa = 11.19NGVHH374 pKa = 7.05LDD376 pKa = 3.67PSDD379 pKa = 3.64YY380 pKa = 10.74TATDD384 pKa = 3.55GNNLVLDD391 pKa = 4.39VACSLNDD398 pKa = 3.62EE399 pKa = 4.53IYY401 pKa = 10.33ILTFGTFSLASISASAITSNTLGVARR427 pKa = 11.84GGTGKK432 pKa = 8.18TVSDD436 pKa = 4.53LSGQAGKK443 pKa = 10.68VLTVNASQNGYY454 pKa = 9.99DD455 pKa = 3.71LASASSAEE463 pKa = 4.25VYY465 pKa = 10.72GFEE468 pKa = 4.55QYY470 pKa = 11.33YY471 pKa = 10.61NPSTLIKK478 pKa = 9.33TVTVVGGKK486 pKa = 8.88FVIDD490 pKa = 4.03GVSQDD495 pKa = 3.43TLEE498 pKa = 4.46LYY500 pKa = 10.19EE501 pKa = 4.37GNTYY505 pKa = 9.74VFNYY509 pKa = 9.27PSSHH513 pKa = 6.56PFKK516 pKa = 10.86FSTTSDD522 pKa = 3.38GTHH525 pKa = 6.58GGGSEE530 pKa = 4.15YY531 pKa = 8.44TTGVTHH537 pKa = 7.27NSSTQVTIVVASNAPTLYY555 pKa = 10.12YY556 pKa = 10.77YY557 pKa = 10.4CASHH561 pKa = 6.75SAMGGQANTPVPADD575 pKa = 3.18NHH577 pKa = 5.92LRR579 pKa = 11.84IITTNQGADD588 pKa = 3.66NISSSQYY595 pKa = 11.63ANFDD599 pKa = 3.4DD600 pKa = 5.36SMFSASGFSFAITNGEE616 pKa = 4.32LIATII621 pKa = 4.25
MM1 pKa = 7.74SYY3 pKa = 10.43LAQVSYY9 pKa = 10.36TGDD12 pKa = 3.32GSTTQYY18 pKa = 11.16SITFPFLDD26 pKa = 3.73STHH29 pKa = 5.29VKK31 pKa = 10.74AFIGGVATTAFTISSSTLTFTSAPANSAVIRR62 pKa = 11.84IEE64 pKa = 3.86RR65 pKa = 11.84QTPTSSRR72 pKa = 11.84LVDD75 pKa = 4.11FTDD78 pKa = 4.41GSVLTEE84 pKa = 3.91SDD86 pKa = 3.6LDD88 pKa = 3.76RR89 pKa = 11.84SADD92 pKa = 3.47QNFYY96 pKa = 10.3IAQEE100 pKa = 3.83ISDD103 pKa = 4.29ANQSAMTIGTDD114 pKa = 4.3DD115 pKa = 4.6KK116 pKa = 11.5FDD118 pKa = 3.85AQSKK122 pKa = 9.01IIKK125 pKa = 9.87NVANPVDD132 pKa = 3.74NQDD135 pKa = 3.81AVTKK139 pKa = 10.51YY140 pKa = 10.39YY141 pKa = 11.04LEE143 pKa = 4.28NTWLSSANKK152 pKa = 8.63TALTTVNSNITAITNVNSNSTNINAVNSNSTNINLVAGSIGSVNTVATDD201 pKa = 3.31IAKK204 pKa = 10.49VIAVANDD211 pKa = 3.2LAEE214 pKa = 4.27AVSEE218 pKa = 4.53VEE220 pKa = 4.57TVANDD225 pKa = 3.74LNEE228 pKa = 4.33STSEE232 pKa = 3.68IDD234 pKa = 3.35TVANAITNVDD244 pKa = 3.0IVGNAIANVNTLAGVSNLQNLANAHH269 pKa = 5.0SQIVTVGNNIPAVEE283 pKa = 4.14AFGNTYY289 pKa = 9.96KK290 pKa = 10.42ISASAPSSPIEE301 pKa = 3.8GTLWFDD307 pKa = 3.32TTADD311 pKa = 3.03IMKK314 pKa = 9.86VYY316 pKa = 10.5DD317 pKa = 3.6GSSFVNAGSSVNGTSGRR334 pKa = 11.84FKK336 pKa = 11.11YY337 pKa = 10.11IATAGQTTFTGASHH351 pKa = 7.24SDD353 pKa = 3.16TGSNTLTYY361 pKa = 10.48DD362 pKa = 2.98AGFIDD367 pKa = 4.02VFKK370 pKa = 11.19NGVHH374 pKa = 7.05LDD376 pKa = 3.67PSDD379 pKa = 3.64YY380 pKa = 10.74TATDD384 pKa = 3.55GNNLVLDD391 pKa = 4.39VACSLNDD398 pKa = 3.62EE399 pKa = 4.53IYY401 pKa = 10.33ILTFGTFSLASISASAITSNTLGVARR427 pKa = 11.84GGTGKK432 pKa = 8.18TVSDD436 pKa = 4.53LSGQAGKK443 pKa = 10.68VLTVNASQNGYY454 pKa = 9.99DD455 pKa = 3.71LASASSAEE463 pKa = 4.25VYY465 pKa = 10.72GFEE468 pKa = 4.55QYY470 pKa = 11.33YY471 pKa = 10.61NPSTLIKK478 pKa = 9.33TVTVVGGKK486 pKa = 8.88FVIDD490 pKa = 4.03GVSQDD495 pKa = 3.43TLEE498 pKa = 4.46LYY500 pKa = 10.19EE501 pKa = 4.37GNTYY505 pKa = 9.74VFNYY509 pKa = 9.27PSSHH513 pKa = 6.56PFKK516 pKa = 10.86FSTTSDD522 pKa = 3.38GTHH525 pKa = 6.58GGGSEE530 pKa = 4.15YY531 pKa = 8.44TTGVTHH537 pKa = 7.27NSSTQVTIVVASNAPTLYY555 pKa = 10.12YY556 pKa = 10.77YY557 pKa = 10.4CASHH561 pKa = 6.75SAMGGQANTPVPADD575 pKa = 3.18NHH577 pKa = 5.92LRR579 pKa = 11.84IITTNQGADD588 pKa = 3.66NISSSQYY595 pKa = 11.63ANFDD599 pKa = 3.4DD600 pKa = 5.36SMFSASGFSFAITNGEE616 pKa = 4.32LIATII621 pKa = 4.25
Molecular weight: 64.91 kDa
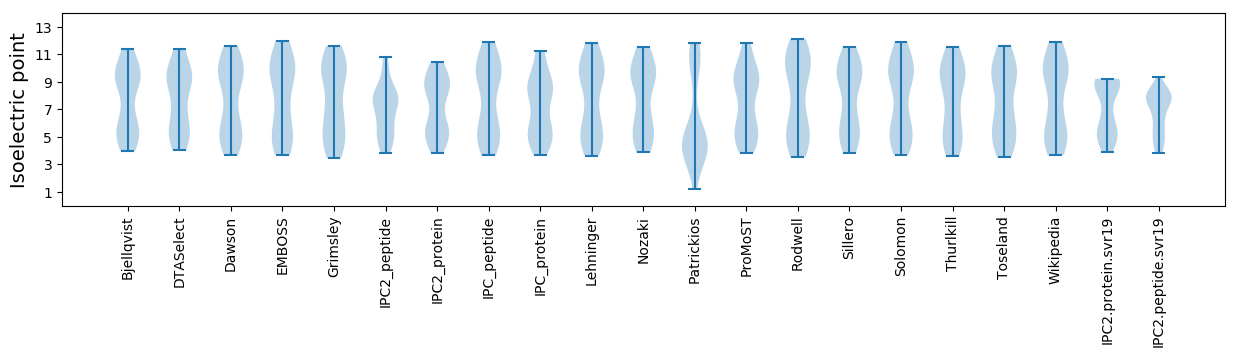
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6S4PDT1|A0A6S4PDT1_9CAUD Uncharacterized protein OS=uncultured phage_MedDCM-OCT-S30-C28 OX=2741076 PE=4 SV=1
MM1 pKa = 7.48KK2 pKa = 10.47GAINSLNNRR11 pKa = 11.84GAMVEE16 pKa = 4.3LVDD19 pKa = 4.32TPVLGTGLLRR29 pKa = 11.84GEE31 pKa = 4.55GSSPFRR37 pKa = 11.84PTKK40 pKa = 10.39DD41 pKa = 4.11DD42 pKa = 3.54YY43 pKa = 11.99DD44 pKa = 3.67GVYY47 pKa = 10.32DD48 pKa = 3.95ISYY51 pKa = 10.75SSTIEE56 pKa = 3.9KK57 pKa = 10.54KK58 pKa = 10.65KK59 pKa = 10.87GVVNNKK65 pKa = 9.37KK66 pKa = 10.85GNFGSLRR73 pKa = 11.84KK74 pKa = 9.58NHH76 pKa = 5.88TFHH79 pKa = 7.53LNNTSANNSLSTRR92 pKa = 11.84CFLKK96 pKa = 10.82LISSTPVWWSSRR108 pKa = 11.84RR109 pKa = 11.84FTISTRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84TPVLVSTKK125 pKa = 10.47EE126 pKa = 3.9MFSQCPQKK134 pKa = 10.57LIKK137 pKa = 9.98HH138 pKa = 6.25HH139 pKa = 6.0YY140 pKa = 9.45SYY142 pKa = 11.9NN143 pKa = 3.43
MM1 pKa = 7.48KK2 pKa = 10.47GAINSLNNRR11 pKa = 11.84GAMVEE16 pKa = 4.3LVDD19 pKa = 4.32TPVLGTGLLRR29 pKa = 11.84GEE31 pKa = 4.55GSSPFRR37 pKa = 11.84PTKK40 pKa = 10.39DD41 pKa = 4.11DD42 pKa = 3.54YY43 pKa = 11.99DD44 pKa = 3.67GVYY47 pKa = 10.32DD48 pKa = 3.95ISYY51 pKa = 10.75SSTIEE56 pKa = 3.9KK57 pKa = 10.54KK58 pKa = 10.65KK59 pKa = 10.87GVVNNKK65 pKa = 9.37KK66 pKa = 10.85GNFGSLRR73 pKa = 11.84KK74 pKa = 9.58NHH76 pKa = 5.88TFHH79 pKa = 7.53LNNTSANNSLSTRR92 pKa = 11.84CFLKK96 pKa = 10.82LISSTPVWWSSRR108 pKa = 11.84RR109 pKa = 11.84FTISTRR115 pKa = 11.84RR116 pKa = 11.84RR117 pKa = 11.84TPVLVSTKK125 pKa = 10.47EE126 pKa = 3.9MFSQCPQKK134 pKa = 10.57LIKK137 pKa = 9.98HH138 pKa = 6.25HH139 pKa = 6.0YY140 pKa = 9.45SYY142 pKa = 11.9NN143 pKa = 3.43
Molecular weight: 16.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11730 |
40 |
1231 |
230.0 |
25.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.556 ± 0.384 | 0.878 ± 0.161 |
6.01 ± 0.248 | 6.428 ± 0.495 |
4.263 ± 0.314 | 6.044 ± 0.413 |
1.628 ± 0.237 | 6.667 ± 0.274 |
8.986 ± 0.639 | 7.945 ± 0.363 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.114 ± 0.186 | 6.394 ± 0.282 |
3.214 ± 0.258 | 4.211 ± 0.24 |
4.177 ± 0.279 | 7.238 ± 0.536 |
6.726 ± 0.541 | 5.968 ± 0.317 |
0.929 ± 0.143 | 3.623 ± 0.152 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
