
Pseudogymnoascus sp. VKM F-4516 (FW-969)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes; Leotiomycetes incertae sedis; Pseudeurotiaceae; Pseudogymnoascus; unclassified Pseudogymnoascus
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
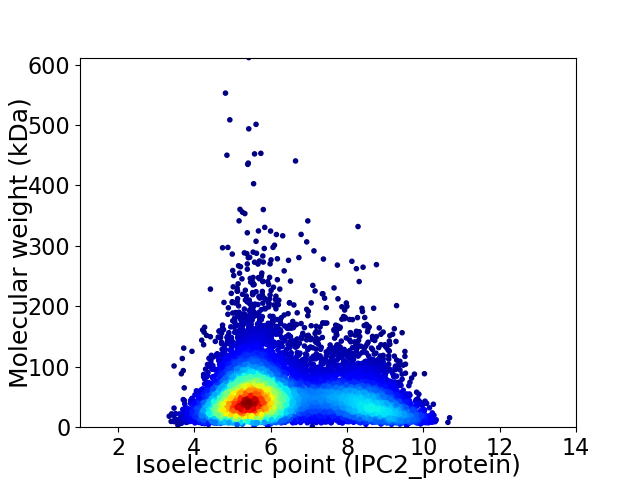
Virtual 2D-PAGE plot for 9362 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
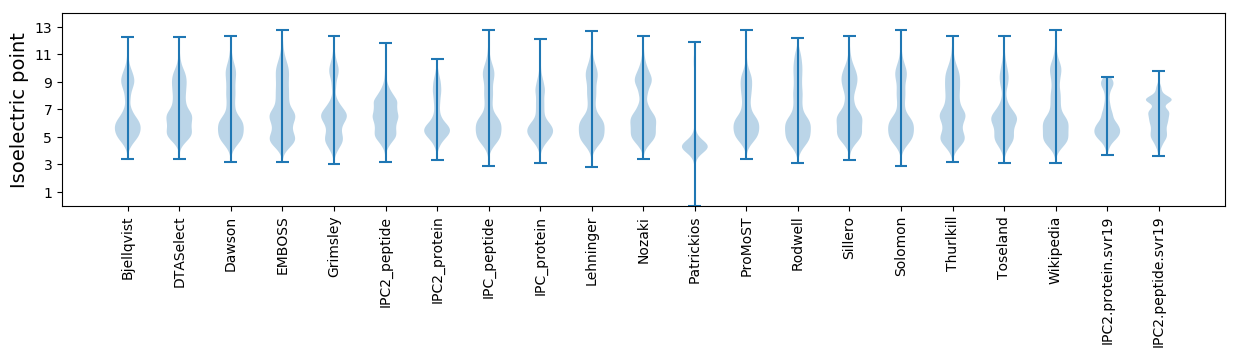
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A094DBN8|A0A094DBN8_9PEZI TBPIP domain-containing protein OS=Pseudogymnoascus sp. VKM F-4516 (FW-969) OX=1420910 GN=V497_02805 PE=3 SV=1
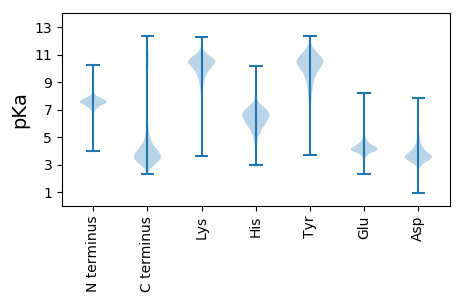
MM1 pKa = 7.54SGYY4 pKa = 11.32GNDD7 pKa = 4.3NNDD10 pKa = 3.09SSNTRR15 pKa = 11.84GGDD18 pKa = 3.31SYY20 pKa = 12.02GSGNTGSGNNDD31 pKa = 3.02SYY33 pKa = 11.79GSSGNTGSDD42 pKa = 3.44SYY44 pKa = 11.59GSSGNTRR51 pKa = 11.84SSGNDD56 pKa = 3.14DD57 pKa = 4.01SYY59 pKa = 11.97GSSNTRR65 pKa = 11.84SSDD68 pKa = 3.38NNDD71 pKa = 3.1SYY73 pKa = 11.93GSSNTRR79 pKa = 11.84SSDD82 pKa = 3.38NNDD85 pKa = 3.1SYY87 pKa = 11.93GSSNTRR93 pKa = 11.84SSDD96 pKa = 3.38NNDD99 pKa = 3.16SYY101 pKa = 11.89GSSGNSRR108 pKa = 11.84SNDD111 pKa = 3.05NDD113 pKa = 3.82SYY115 pKa = 11.64GSSNKK120 pKa = 9.81SSSDD124 pKa = 3.34NNDD127 pKa = 2.94SYY129 pKa = 11.93GSSNTRR135 pKa = 11.84SGDD138 pKa = 3.24NDD140 pKa = 4.0SYY142 pKa = 11.66GSSGNSRR149 pKa = 11.84SDD151 pKa = 3.38NNDD154 pKa = 2.75SYY156 pKa = 11.99GSSNKK161 pKa = 9.78SSSGNNDD168 pKa = 2.79SYY170 pKa = 11.87GSSNNDD176 pKa = 2.76SYY178 pKa = 11.96GSSNTRR184 pKa = 11.84SSGNDD189 pKa = 3.14DD190 pKa = 4.32SYY192 pKa = 12.16GSSNNNNSSDD202 pKa = 3.85SYY204 pKa = 11.03GSSGGKK210 pKa = 7.85TGNSTVDD217 pKa = 3.35KK218 pKa = 10.63LVDD221 pKa = 3.39KK222 pKa = 11.21ASDD225 pKa = 3.61FLGSNNSSSGNNNSNYY241 pKa = 10.73
MM1 pKa = 7.54SGYY4 pKa = 11.32GNDD7 pKa = 4.3NNDD10 pKa = 3.09SSNTRR15 pKa = 11.84GGDD18 pKa = 3.31SYY20 pKa = 12.02GSGNTGSGNNDD31 pKa = 3.02SYY33 pKa = 11.79GSSGNTGSDD42 pKa = 3.44SYY44 pKa = 11.59GSSGNTRR51 pKa = 11.84SSGNDD56 pKa = 3.14DD57 pKa = 4.01SYY59 pKa = 11.97GSSNTRR65 pKa = 11.84SSDD68 pKa = 3.38NNDD71 pKa = 3.1SYY73 pKa = 11.93GSSNTRR79 pKa = 11.84SSDD82 pKa = 3.38NNDD85 pKa = 3.1SYY87 pKa = 11.93GSSNTRR93 pKa = 11.84SSDD96 pKa = 3.38NNDD99 pKa = 3.16SYY101 pKa = 11.89GSSGNSRR108 pKa = 11.84SNDD111 pKa = 3.05NDD113 pKa = 3.82SYY115 pKa = 11.64GSSNKK120 pKa = 9.81SSSDD124 pKa = 3.34NNDD127 pKa = 2.94SYY129 pKa = 11.93GSSNTRR135 pKa = 11.84SGDD138 pKa = 3.24NDD140 pKa = 4.0SYY142 pKa = 11.66GSSGNSRR149 pKa = 11.84SDD151 pKa = 3.38NNDD154 pKa = 2.75SYY156 pKa = 11.99GSSNKK161 pKa = 9.78SSSGNNDD168 pKa = 2.79SYY170 pKa = 11.87GSSNNDD176 pKa = 2.76SYY178 pKa = 11.96GSSNTRR184 pKa = 11.84SSGNDD189 pKa = 3.14DD190 pKa = 4.32SYY192 pKa = 12.16GSSNNNNSSDD202 pKa = 3.85SYY204 pKa = 11.03GSSGGKK210 pKa = 7.85TGNSTVDD217 pKa = 3.35KK218 pKa = 10.63LVDD221 pKa = 3.39KK222 pKa = 11.21ASDD225 pKa = 3.61FLGSNNSSSGNNNSNYY241 pKa = 10.73
Molecular weight: 24.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A094E0U6|A0A094E0U6_9PEZI Arf-GAP domain-containing protein (Fragment) OS=Pseudogymnoascus sp. VKM F-4516 (FW-969) OX=1420910 GN=V497_01425 PE=4 SV=1
MM1 pKa = 6.8ITQRR5 pKa = 11.84QATASLCRR13 pKa = 11.84KK14 pKa = 9.33RR15 pKa = 11.84NTVFVTLRR23 pKa = 11.84LRR25 pKa = 11.84VKK27 pKa = 10.4SPIRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84LGEE37 pKa = 4.03SFGDD41 pKa = 3.74TVGGGGHH48 pKa = 5.88EE49 pKa = 4.13RR50 pKa = 11.84WAGGPGRR57 pKa = 11.84CSQGVLGRR65 pKa = 11.84ARR67 pKa = 11.84RR68 pKa = 11.84FGRR71 pKa = 11.84GRR73 pKa = 11.84WGWGRR78 pKa = 11.84LEE80 pKa = 3.97MGTWEE85 pKa = 4.5SFPGVTPGGFGVGFGLLVRR104 pKa = 11.84FDD106 pKa = 4.55GSFLCLDD113 pKa = 4.32GGRR116 pKa = 11.84EE117 pKa = 4.16GGAGEE122 pKa = 4.38SGNVAGILLVRR133 pKa = 11.84KK134 pKa = 9.68VKK136 pKa = 10.66LGQRR140 pKa = 11.84DD141 pKa = 2.76AWIPHH146 pKa = 5.56TLPVGRR152 pKa = 11.84RR153 pKa = 11.84DD154 pKa = 3.89ADD156 pKa = 3.54EE157 pKa = 5.72LSDD160 pKa = 3.65KK161 pKa = 10.5RR162 pKa = 11.84CPIGGDD168 pKa = 2.99IWCGGINAVPIKK180 pKa = 8.33TAVMRR185 pKa = 11.84APDD188 pKa = 3.67LAYY191 pKa = 10.4VFRR194 pKa = 11.84PAVTSMPVHH203 pKa = 5.17QLKK206 pKa = 10.31PRR208 pKa = 11.84GIAVTPGNFFQALSVDD224 pKa = 3.88MEE226 pKa = 4.36HH227 pKa = 7.22AGNSVYY233 pKa = 10.74DD234 pKa = 4.14LRR236 pKa = 11.84GQPTWTSSVTEE247 pKa = 4.23NVDD250 pKa = 2.87CDD252 pKa = 3.66MPNFSTPRR260 pKa = 11.84TSFNN264 pKa = 3.31
MM1 pKa = 6.8ITQRR5 pKa = 11.84QATASLCRR13 pKa = 11.84KK14 pKa = 9.33RR15 pKa = 11.84NTVFVTLRR23 pKa = 11.84LRR25 pKa = 11.84VKK27 pKa = 10.4SPIRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84LGEE37 pKa = 4.03SFGDD41 pKa = 3.74TVGGGGHH48 pKa = 5.88EE49 pKa = 4.13RR50 pKa = 11.84WAGGPGRR57 pKa = 11.84CSQGVLGRR65 pKa = 11.84ARR67 pKa = 11.84RR68 pKa = 11.84FGRR71 pKa = 11.84GRR73 pKa = 11.84WGWGRR78 pKa = 11.84LEE80 pKa = 3.97MGTWEE85 pKa = 4.5SFPGVTPGGFGVGFGLLVRR104 pKa = 11.84FDD106 pKa = 4.55GSFLCLDD113 pKa = 4.32GGRR116 pKa = 11.84EE117 pKa = 4.16GGAGEE122 pKa = 4.38SGNVAGILLVRR133 pKa = 11.84KK134 pKa = 9.68VKK136 pKa = 10.66LGQRR140 pKa = 11.84DD141 pKa = 2.76AWIPHH146 pKa = 5.56TLPVGRR152 pKa = 11.84RR153 pKa = 11.84DD154 pKa = 3.89ADD156 pKa = 3.54EE157 pKa = 5.72LSDD160 pKa = 3.65KK161 pKa = 10.5RR162 pKa = 11.84CPIGGDD168 pKa = 2.99IWCGGINAVPIKK180 pKa = 8.33TAVMRR185 pKa = 11.84APDD188 pKa = 3.67LAYY191 pKa = 10.4VFRR194 pKa = 11.84PAVTSMPVHH203 pKa = 5.17QLKK206 pKa = 10.31PRR208 pKa = 11.84GIAVTPGNFFQALSVDD224 pKa = 3.88MEE226 pKa = 4.36HH227 pKa = 7.22AGNSVYY233 pKa = 10.74DD234 pKa = 4.14LRR236 pKa = 11.84GQPTWTSSVTEE247 pKa = 4.23NVDD250 pKa = 2.87CDD252 pKa = 3.66MPNFSTPRR260 pKa = 11.84TSFNN264 pKa = 3.31
Molecular weight: 28.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4879565 |
8 |
5496 |
521.2 |
57.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.016 ± 0.027 | 1.171 ± 0.01 |
5.634 ± 0.015 | 6.304 ± 0.024 |
3.608 ± 0.015 | 7.266 ± 0.026 |
2.224 ± 0.01 | 5.0 ± 0.018 |
5.042 ± 0.022 | 8.67 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.137 ± 0.01 | 3.735 ± 0.014 |
6.01 ± 0.025 | 3.854 ± 0.019 |
5.819 ± 0.02 | 8.141 ± 0.025 |
6.071 ± 0.018 | 6.131 ± 0.019 |
1.406 ± 0.008 | 2.763 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
