
Citrus psorosis virus (isolate Spain/P-121) (CPsV) (Citrus ringspot virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Milneviricetes; Serpentovirales; Aspiviridae; Ophiovirus; Citrus psorosis ophiovirus
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
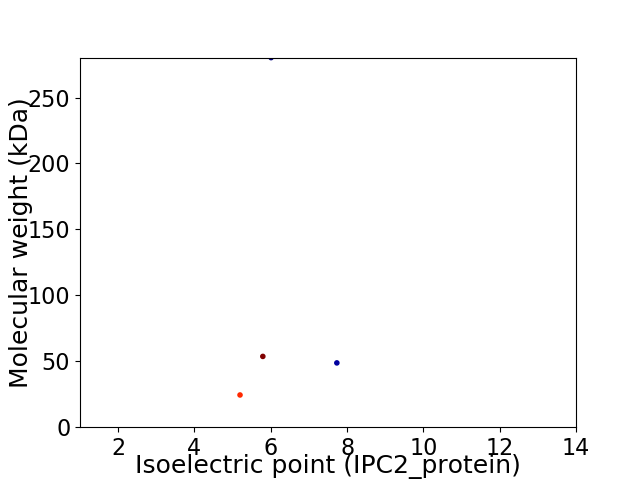
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
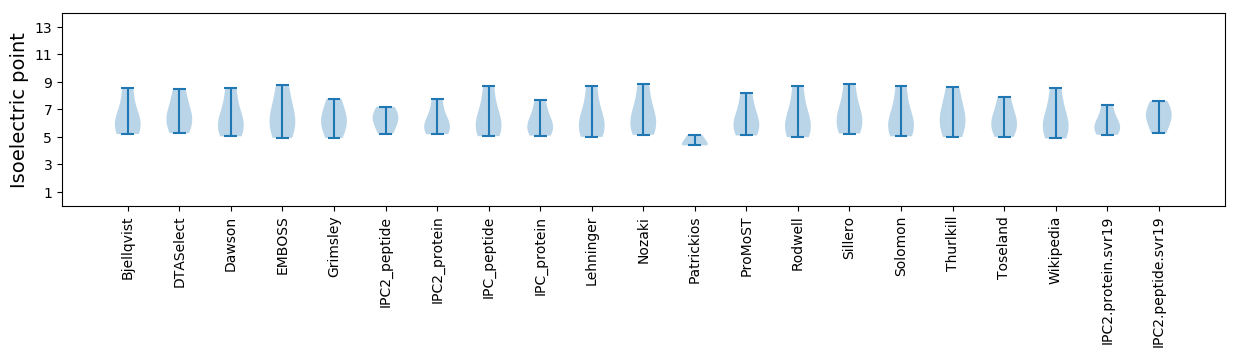
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q6DN67|L_CPSVP RNA-directed RNA polymerase L OS=Citrus psorosis virus (isolate Spain/P-121) OX=652963 GN=L PE=3 SV=1
MM1 pKa = 7.69AEE3 pKa = 4.06YY4 pKa = 10.47IEE6 pKa = 4.14VRR8 pKa = 11.84VEE10 pKa = 4.27NLHH13 pKa = 6.35KK14 pKa = 10.21WGLEE18 pKa = 3.48INMEE22 pKa = 4.47RR23 pKa = 11.84IEE25 pKa = 4.28KK26 pKa = 10.09LSKK29 pKa = 9.85RR30 pKa = 11.84LKK32 pKa = 10.66SIVDD36 pKa = 3.54EE37 pKa = 4.3DD38 pKa = 5.28CIMKK42 pKa = 9.32TSRR45 pKa = 11.84IIGIWMFMPEE55 pKa = 4.06IVQEE59 pKa = 4.21SLKK62 pKa = 10.76DD63 pKa = 3.79SPLMTQKK70 pKa = 11.06AWIIPHH76 pKa = 6.12EE77 pKa = 4.36KK78 pKa = 9.32TYY80 pKa = 9.21KK81 pKa = 9.68TIYY84 pKa = 10.14GKK86 pKa = 10.31DD87 pKa = 3.85GIQMAVTQNEE97 pKa = 3.65EE98 pKa = 4.37DD99 pKa = 3.62LFKK102 pKa = 11.07DD103 pKa = 3.46SEE105 pKa = 4.26FFMISRR111 pKa = 11.84CDD113 pKa = 3.46SVMLTKK119 pKa = 10.7NNKK122 pKa = 8.8TIILNKK128 pKa = 10.14EE129 pKa = 3.99LLNCNMSEE137 pKa = 4.27DD138 pKa = 3.8MLFNMLSCQEE148 pKa = 3.76QDD150 pKa = 2.85ITEE153 pKa = 4.04EE154 pKa = 3.99LMKK157 pKa = 10.7KK158 pKa = 8.73MKK160 pKa = 9.96TIISSNPKK168 pKa = 9.57EE169 pKa = 3.97RR170 pKa = 11.84LEE172 pKa = 4.6DD173 pKa = 3.34KK174 pKa = 10.6TEE176 pKa = 3.84EE177 pKa = 4.23VFWNSTRR184 pKa = 11.84ILNWIQHH191 pKa = 5.63NDD193 pKa = 3.09NSRR196 pKa = 11.84SNSSDD201 pKa = 2.86NSFRR205 pKa = 11.84EE206 pKa = 4.1
MM1 pKa = 7.69AEE3 pKa = 4.06YY4 pKa = 10.47IEE6 pKa = 4.14VRR8 pKa = 11.84VEE10 pKa = 4.27NLHH13 pKa = 6.35KK14 pKa = 10.21WGLEE18 pKa = 3.48INMEE22 pKa = 4.47RR23 pKa = 11.84IEE25 pKa = 4.28KK26 pKa = 10.09LSKK29 pKa = 9.85RR30 pKa = 11.84LKK32 pKa = 10.66SIVDD36 pKa = 3.54EE37 pKa = 4.3DD38 pKa = 5.28CIMKK42 pKa = 9.32TSRR45 pKa = 11.84IIGIWMFMPEE55 pKa = 4.06IVQEE59 pKa = 4.21SLKK62 pKa = 10.76DD63 pKa = 3.79SPLMTQKK70 pKa = 11.06AWIIPHH76 pKa = 6.12EE77 pKa = 4.36KK78 pKa = 9.32TYY80 pKa = 9.21KK81 pKa = 9.68TIYY84 pKa = 10.14GKK86 pKa = 10.31DD87 pKa = 3.85GIQMAVTQNEE97 pKa = 3.65EE98 pKa = 4.37DD99 pKa = 3.62LFKK102 pKa = 11.07DD103 pKa = 3.46SEE105 pKa = 4.26FFMISRR111 pKa = 11.84CDD113 pKa = 3.46SVMLTKK119 pKa = 10.7NNKK122 pKa = 8.8TIILNKK128 pKa = 10.14EE129 pKa = 3.99LLNCNMSEE137 pKa = 4.27DD138 pKa = 3.8MLFNMLSCQEE148 pKa = 3.76QDD150 pKa = 2.85ITEE153 pKa = 4.04EE154 pKa = 3.99LMKK157 pKa = 10.7KK158 pKa = 8.73MKK160 pKa = 9.96TIISSNPKK168 pKa = 9.57EE169 pKa = 3.97RR170 pKa = 11.84LEE172 pKa = 4.6DD173 pKa = 3.34KK174 pKa = 10.6TEE176 pKa = 3.84EE177 pKa = 4.23VFWNSTRR184 pKa = 11.84ILNWIQHH191 pKa = 5.63NDD193 pKa = 3.09NSRR196 pKa = 11.84SNSSDD201 pKa = 2.86NSFRR205 pKa = 11.84EE206 pKa = 4.1
Molecular weight: 24.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q6DN65|VP54_CPSVP Uncharacterized 54 kDa protein OS=Citrus psorosis virus (isolate Spain/P-121) OX=652963 GN=54K PE=4 SV=1
MM1 pKa = 7.81SIPIKK6 pKa = 10.54VSQLIDD12 pKa = 3.72AKK14 pKa = 9.77TKK16 pKa = 9.6WEE18 pKa = 4.07SEE20 pKa = 4.29KK21 pKa = 10.71EE22 pKa = 3.93KK23 pKa = 11.15SPAEE27 pKa = 3.77ILTLLKK33 pKa = 10.05TYY35 pKa = 10.58GVLRR39 pKa = 11.84DD40 pKa = 3.71TGALSSKK47 pKa = 9.24STAFLSGLSKK57 pKa = 10.81DD58 pKa = 3.48KK59 pKa = 10.97RR60 pKa = 11.84VILTDD65 pKa = 3.1EE66 pKa = 4.37GLAEE70 pKa = 4.47FKK72 pKa = 10.84SVEE75 pKa = 4.09NPHH78 pKa = 6.81DD79 pKa = 3.78QGEE82 pKa = 4.54KK83 pKa = 9.89PAFTLASSSSIKK95 pKa = 8.8ITNIEE100 pKa = 4.12VIKK103 pKa = 10.76QKK105 pKa = 10.04MEE107 pKa = 3.8QEE109 pKa = 4.4TFQIDD114 pKa = 3.56QLKK117 pKa = 10.62LKK119 pKa = 9.79EE120 pKa = 4.02QIEE123 pKa = 4.34NFIKK127 pKa = 10.02TVTLDD132 pKa = 3.54DD133 pKa = 3.75EE134 pKa = 5.54SYY136 pKa = 10.45TEE138 pKa = 4.08GEE140 pKa = 4.09IIIKK144 pKa = 10.52HH145 pKa = 6.1FGNPDD150 pKa = 3.1AEE152 pKa = 4.45LNMLITAGTKK162 pKa = 9.7ILDD165 pKa = 3.61GLVYY169 pKa = 10.93VSMKK173 pKa = 10.87GDD175 pKa = 3.55TKK177 pKa = 11.19SLNLFKK183 pKa = 10.21MEE185 pKa = 4.27QVDD188 pKa = 4.77GVCDD192 pKa = 3.55SDD194 pKa = 3.61IIKK197 pKa = 10.44NIRR200 pKa = 11.84VAKK203 pKa = 10.07RR204 pKa = 11.84AIQAAFVLIFTQGSLPGKK222 pKa = 10.24ADD224 pKa = 4.07DD225 pKa = 3.96KK226 pKa = 11.21RR227 pKa = 11.84KK228 pKa = 10.14VPEE231 pKa = 3.91FVKK234 pKa = 10.82SKK236 pKa = 10.76LYY238 pKa = 11.05DD239 pKa = 3.77GDD241 pKa = 3.79VSLSQISEE249 pKa = 4.34EE250 pKa = 4.38LSHH253 pKa = 6.94APTKK257 pKa = 10.44KK258 pKa = 9.68FPARR262 pKa = 11.84VFLKK266 pKa = 10.2IDD268 pKa = 3.38IDD270 pKa = 3.93NLPSAVCSRR279 pKa = 11.84CKK281 pKa = 10.78LNIAGNRR288 pKa = 11.84SVRR291 pKa = 11.84YY292 pKa = 9.72AGFASSFQTKK302 pKa = 9.28QKK304 pKa = 10.61LSPAVGATPEE314 pKa = 4.09SLMPLLEE321 pKa = 4.29TNQKK325 pKa = 9.31IEE327 pKa = 3.85KK328 pKa = 9.86SIAIRR333 pKa = 11.84DD334 pKa = 3.87FLKK337 pKa = 10.4TMEE340 pKa = 4.63GQWKK344 pKa = 7.65NQKK347 pKa = 9.56RR348 pKa = 11.84LHH350 pKa = 6.31PLSDD354 pKa = 3.68EE355 pKa = 4.28KK356 pKa = 10.79PTIKK360 pKa = 10.7NFTLKK365 pKa = 9.16LTCAIIYY372 pKa = 10.26SLTPDD377 pKa = 3.3GRR379 pKa = 11.84IDD381 pKa = 3.25MAEE384 pKa = 4.19RR385 pKa = 11.84IITDD389 pKa = 3.52KK390 pKa = 11.58NKK392 pKa = 10.24GFQNDD397 pKa = 3.15RR398 pKa = 11.84NFFGDD403 pKa = 3.64GEE405 pKa = 4.97GPTRR409 pKa = 11.84TWSVLTKK416 pKa = 10.45PEE418 pKa = 4.31ADD420 pKa = 3.62FSNITVDD427 pKa = 3.37GLKK430 pKa = 10.84GIFGVASTMM439 pKa = 3.81
MM1 pKa = 7.81SIPIKK6 pKa = 10.54VSQLIDD12 pKa = 3.72AKK14 pKa = 9.77TKK16 pKa = 9.6WEE18 pKa = 4.07SEE20 pKa = 4.29KK21 pKa = 10.71EE22 pKa = 3.93KK23 pKa = 11.15SPAEE27 pKa = 3.77ILTLLKK33 pKa = 10.05TYY35 pKa = 10.58GVLRR39 pKa = 11.84DD40 pKa = 3.71TGALSSKK47 pKa = 9.24STAFLSGLSKK57 pKa = 10.81DD58 pKa = 3.48KK59 pKa = 10.97RR60 pKa = 11.84VILTDD65 pKa = 3.1EE66 pKa = 4.37GLAEE70 pKa = 4.47FKK72 pKa = 10.84SVEE75 pKa = 4.09NPHH78 pKa = 6.81DD79 pKa = 3.78QGEE82 pKa = 4.54KK83 pKa = 9.89PAFTLASSSSIKK95 pKa = 8.8ITNIEE100 pKa = 4.12VIKK103 pKa = 10.76QKK105 pKa = 10.04MEE107 pKa = 3.8QEE109 pKa = 4.4TFQIDD114 pKa = 3.56QLKK117 pKa = 10.62LKK119 pKa = 9.79EE120 pKa = 4.02QIEE123 pKa = 4.34NFIKK127 pKa = 10.02TVTLDD132 pKa = 3.54DD133 pKa = 3.75EE134 pKa = 5.54SYY136 pKa = 10.45TEE138 pKa = 4.08GEE140 pKa = 4.09IIIKK144 pKa = 10.52HH145 pKa = 6.1FGNPDD150 pKa = 3.1AEE152 pKa = 4.45LNMLITAGTKK162 pKa = 9.7ILDD165 pKa = 3.61GLVYY169 pKa = 10.93VSMKK173 pKa = 10.87GDD175 pKa = 3.55TKK177 pKa = 11.19SLNLFKK183 pKa = 10.21MEE185 pKa = 4.27QVDD188 pKa = 4.77GVCDD192 pKa = 3.55SDD194 pKa = 3.61IIKK197 pKa = 10.44NIRR200 pKa = 11.84VAKK203 pKa = 10.07RR204 pKa = 11.84AIQAAFVLIFTQGSLPGKK222 pKa = 10.24ADD224 pKa = 4.07DD225 pKa = 3.96KK226 pKa = 11.21RR227 pKa = 11.84KK228 pKa = 10.14VPEE231 pKa = 3.91FVKK234 pKa = 10.82SKK236 pKa = 10.76LYY238 pKa = 11.05DD239 pKa = 3.77GDD241 pKa = 3.79VSLSQISEE249 pKa = 4.34EE250 pKa = 4.38LSHH253 pKa = 6.94APTKK257 pKa = 10.44KK258 pKa = 9.68FPARR262 pKa = 11.84VFLKK266 pKa = 10.2IDD268 pKa = 3.38IDD270 pKa = 3.93NLPSAVCSRR279 pKa = 11.84CKK281 pKa = 10.78LNIAGNRR288 pKa = 11.84SVRR291 pKa = 11.84YY292 pKa = 9.72AGFASSFQTKK302 pKa = 9.28QKK304 pKa = 10.61LSPAVGATPEE314 pKa = 4.09SLMPLLEE321 pKa = 4.29TNQKK325 pKa = 9.31IEE327 pKa = 3.85KK328 pKa = 9.86SIAIRR333 pKa = 11.84DD334 pKa = 3.87FLKK337 pKa = 10.4TMEE340 pKa = 4.63GQWKK344 pKa = 7.65NQKK347 pKa = 9.56RR348 pKa = 11.84LHH350 pKa = 6.31PLSDD354 pKa = 3.68EE355 pKa = 4.28KK356 pKa = 10.79PTIKK360 pKa = 10.7NFTLKK365 pKa = 9.16LTCAIIYY372 pKa = 10.26SLTPDD377 pKa = 3.3GRR379 pKa = 11.84IDD381 pKa = 3.25MAEE384 pKa = 4.19RR385 pKa = 11.84IITDD389 pKa = 3.52KK390 pKa = 11.58NKK392 pKa = 10.24GFQNDD397 pKa = 3.15RR398 pKa = 11.84NFFGDD403 pKa = 3.64GEE405 pKa = 4.97GPTRR409 pKa = 11.84TWSVLTKK416 pKa = 10.45PEE418 pKa = 4.31ADD420 pKa = 3.62FSNITVDD427 pKa = 3.37GLKK430 pKa = 10.84GIFGVASTMM439 pKa = 3.81
Molecular weight: 48.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3537 |
206 |
2416 |
884.3 |
101.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.76 ± 0.582 | 1.159 ± 0.15 |
5.966 ± 0.38 | 9.019 ± 0.56 |
4.015 ± 0.206 | 4.891 ± 0.438 |
1.414 ± 0.173 | 9.358 ± 0.695 |
9.867 ± 0.18 | 9.075 ± 0.512 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.902 ± 0.473 | 5.909 ± 0.581 |
2.29 ± 0.458 | 2.771 ± 0.366 |
4.467 ± 0.243 | 7.888 ± 0.599 |
5.146 ± 0.462 | 4.891 ± 0.399 |
0.876 ± 0.197 | 3.308 ± 0.86 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
