
Nitrososphaeraceae archaeon
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; Nitrososphaeria; Nitrososphaerales; Nitrososphaeraceae; unclassified Nitrososphaeraceae
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
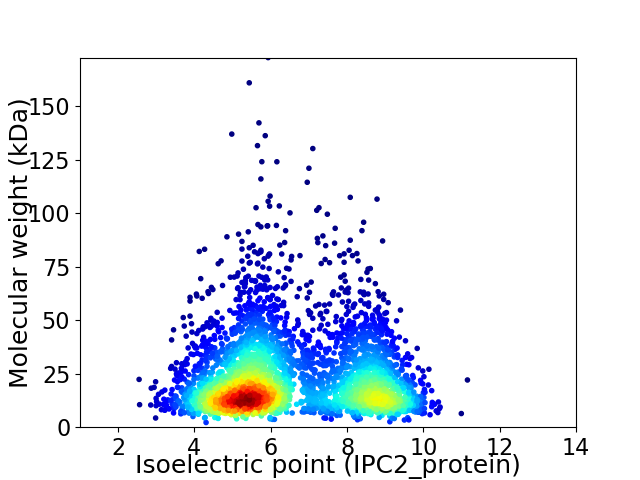
Virtual 2D-PAGE plot for 3773 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6A7LEI3|A0A6A7LEI3_9ARCH Winged helix-turn-helix transcriptional regulator OS=Nitrososphaeraceae archaeon OX=2169593 GN=GEU26_06255 PE=4 SV=1
MM1 pKa = 7.98DD2 pKa = 4.84NKK4 pKa = 10.85SFLISVLGLLLVMVYY19 pKa = 8.81FNPFGLHH26 pKa = 5.96DD27 pKa = 3.88QNVFGSVSDD36 pKa = 5.0DD37 pKa = 3.37EE38 pKa = 5.51DD39 pKa = 4.3NSVTEE44 pKa = 4.5QPLQQPLVNNSTSGIATSIYY64 pKa = 8.39DD65 pKa = 3.36THH67 pKa = 6.49EE68 pKa = 4.32FNVGDD73 pKa = 3.78GAQNLFILIPNEE85 pKa = 3.84GHH87 pKa = 6.65HH88 pKa = 6.76GPGEE92 pKa = 4.01EE93 pKa = 5.04DD94 pKa = 3.09EE95 pKa = 5.28ARR97 pKa = 11.84YY98 pKa = 10.0LNQPFVPEE106 pKa = 4.46SVTISPGTNVVWFNGDD122 pKa = 2.83VGHH125 pKa = 6.15EE126 pKa = 4.18HH127 pKa = 6.69NLAISGGGGSSNPLYY142 pKa = 9.59QTGEE146 pKa = 3.76FSEE149 pKa = 4.51FDD151 pKa = 3.12ARR153 pKa = 11.84NYY155 pKa = 8.45TFNEE159 pKa = 3.84IGDD162 pKa = 3.84FSYY165 pKa = 11.77ADD167 pKa = 3.33TVEE170 pKa = 4.17YY171 pKa = 10.85EE172 pKa = 3.8NGFIMRR178 pKa = 11.84GNISVTDD185 pKa = 4.76DD186 pKa = 4.64DD187 pKa = 5.12PSSQQGTTGSNPPTIGLLMVPTEE210 pKa = 5.12DD211 pKa = 4.87LAQHH215 pKa = 5.43TQDD218 pKa = 3.2LQGRR222 pKa = 11.84GFVIDD227 pKa = 3.73STHH230 pKa = 6.18NFPDD234 pKa = 3.41LRR236 pKa = 11.84EE237 pKa = 4.19GDD239 pKa = 3.93EE240 pKa = 3.95QTLVVWEE247 pKa = 4.39APEE250 pKa = 4.34NSDD253 pKa = 4.17ASTIIADD260 pKa = 3.99LAEE263 pKa = 4.37VSQQFPYY270 pKa = 10.86SS271 pKa = 3.42
MM1 pKa = 7.98DD2 pKa = 4.84NKK4 pKa = 10.85SFLISVLGLLLVMVYY19 pKa = 8.81FNPFGLHH26 pKa = 5.96DD27 pKa = 3.88QNVFGSVSDD36 pKa = 5.0DD37 pKa = 3.37EE38 pKa = 5.51DD39 pKa = 4.3NSVTEE44 pKa = 4.5QPLQQPLVNNSTSGIATSIYY64 pKa = 8.39DD65 pKa = 3.36THH67 pKa = 6.49EE68 pKa = 4.32FNVGDD73 pKa = 3.78GAQNLFILIPNEE85 pKa = 3.84GHH87 pKa = 6.65HH88 pKa = 6.76GPGEE92 pKa = 4.01EE93 pKa = 5.04DD94 pKa = 3.09EE95 pKa = 5.28ARR97 pKa = 11.84YY98 pKa = 10.0LNQPFVPEE106 pKa = 4.46SVTISPGTNVVWFNGDD122 pKa = 2.83VGHH125 pKa = 6.15EE126 pKa = 4.18HH127 pKa = 6.69NLAISGGGGSSNPLYY142 pKa = 9.59QTGEE146 pKa = 3.76FSEE149 pKa = 4.51FDD151 pKa = 3.12ARR153 pKa = 11.84NYY155 pKa = 8.45TFNEE159 pKa = 3.84IGDD162 pKa = 3.84FSYY165 pKa = 11.77ADD167 pKa = 3.33TVEE170 pKa = 4.17YY171 pKa = 10.85EE172 pKa = 3.8NGFIMRR178 pKa = 11.84GNISVTDD185 pKa = 4.76DD186 pKa = 4.64DD187 pKa = 5.12PSSQQGTTGSNPPTIGLLMVPTEE210 pKa = 5.12DD211 pKa = 4.87LAQHH215 pKa = 5.43TQDD218 pKa = 3.2LQGRR222 pKa = 11.84GFVIDD227 pKa = 3.73STHH230 pKa = 6.18NFPDD234 pKa = 3.41LRR236 pKa = 11.84EE237 pKa = 4.19GDD239 pKa = 3.93EE240 pKa = 3.95QTLVVWEE247 pKa = 4.39APEE250 pKa = 4.34NSDD253 pKa = 4.17ASTIIADD260 pKa = 3.99LAEE263 pKa = 4.37VSQQFPYY270 pKa = 10.86SS271 pKa = 3.42
Molecular weight: 29.57 kDa
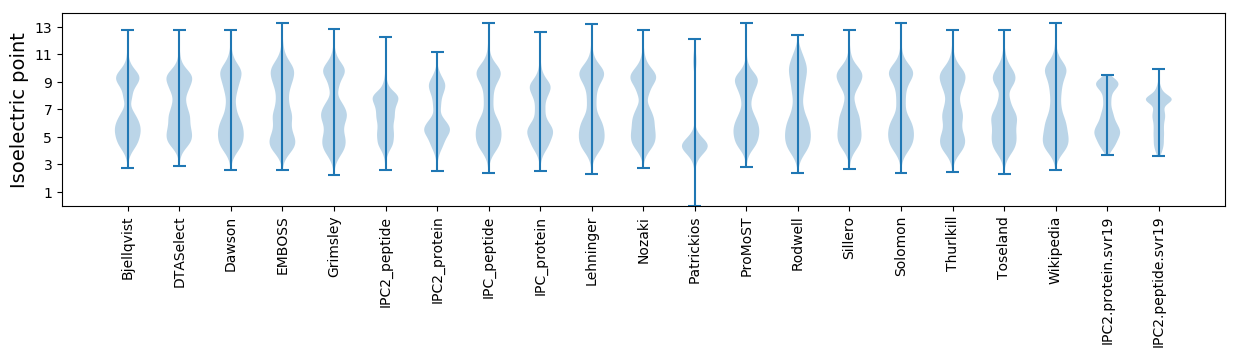
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6A7LM11|A0A6A7LM11_9ARCH Lactamase_B domain-containing protein OS=Nitrososphaeraceae archaeon OX=2169593 GN=GEU26_17090 PE=4 SV=1
MM1 pKa = 7.26LHH3 pKa = 6.72SPLLLLPLPLLRR15 pKa = 11.84PLLLQLLPLLLRR27 pKa = 11.84LLLQLLLLMLRR38 pKa = 11.84PTRR41 pKa = 11.84LLLLLKK47 pKa = 10.48LLQLLLLLLRR57 pKa = 11.84LRR59 pKa = 11.84LLLRR63 pKa = 11.84LLLHH67 pKa = 6.55LLLRR71 pKa = 11.84RR72 pKa = 11.84PLRR75 pKa = 11.84LHH77 pKa = 5.89QLQLLLRR84 pKa = 11.84LLRR87 pKa = 11.84LHH89 pKa = 7.07LKK91 pKa = 10.15LLPLLRR97 pKa = 11.84LNYY100 pKa = 10.11LRR102 pKa = 11.84LRR104 pKa = 11.84LLLPLLLHH112 pKa = 6.74LLPLLLLQIRR122 pKa = 11.84LLLPLLPLLHH132 pKa = 7.31RR133 pKa = 11.84LRR135 pKa = 11.84PLLPLQIRR143 pKa = 11.84QLLPLLPLHH152 pKa = 6.72CLLLLLRR159 pKa = 11.84LLTLHH164 pKa = 6.39QLLPLLPPQSNLMPPPRR181 pKa = 11.84QLLLRR186 pKa = 4.45
MM1 pKa = 7.26LHH3 pKa = 6.72SPLLLLPLPLLRR15 pKa = 11.84PLLLQLLPLLLRR27 pKa = 11.84LLLQLLLLMLRR38 pKa = 11.84PTRR41 pKa = 11.84LLLLLKK47 pKa = 10.48LLQLLLLLLRR57 pKa = 11.84LRR59 pKa = 11.84LLLRR63 pKa = 11.84LLLHH67 pKa = 6.55LLLRR71 pKa = 11.84RR72 pKa = 11.84PLRR75 pKa = 11.84LHH77 pKa = 5.89QLQLLLRR84 pKa = 11.84LLRR87 pKa = 11.84LHH89 pKa = 7.07LKK91 pKa = 10.15LLPLLRR97 pKa = 11.84LNYY100 pKa = 10.11LRR102 pKa = 11.84LRR104 pKa = 11.84LLLPLLLHH112 pKa = 6.74LLPLLLLQIRR122 pKa = 11.84LLLPLLPLLHH132 pKa = 7.31RR133 pKa = 11.84LRR135 pKa = 11.84PLLPLQIRR143 pKa = 11.84QLLPLLPLHH152 pKa = 6.72CLLLLLRR159 pKa = 11.84LLTLHH164 pKa = 6.39QLLPLLPPQSNLMPPPRR181 pKa = 11.84QLLLRR186 pKa = 4.45
Molecular weight: 22.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
824061 |
20 |
1500 |
218.4 |
24.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.164 ± 0.043 | 1.082 ± 0.017 |
5.765 ± 0.031 | 6.414 ± 0.038 |
3.956 ± 0.026 | 6.545 ± 0.041 |
1.951 ± 0.02 | 8.332 ± 0.036 |
6.397 ± 0.041 | 9.111 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.553 ± 0.019 | 5.238 ± 0.035 |
3.826 ± 0.028 | 3.278 ± 0.023 |
5.109 ± 0.037 | 7.873 ± 0.04 |
5.509 ± 0.035 | 6.505 ± 0.034 |
0.93 ± 0.015 | 3.46 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
