
Hypholoma sublateritium (strain FD-334 SS-4)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Strophariaceae; Hypholoma; Hypholoma sublateritium
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
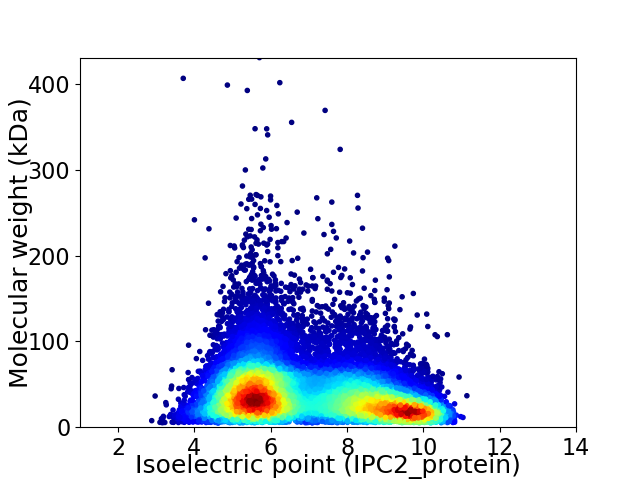
Virtual 2D-PAGE plot for 17638 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0D2KQ62|A0A0D2KQ62_HYPSF Uncharacterized protein OS=Hypholoma sublateritium (strain FD-334 SS-4) OX=945553 GN=HYPSUDRAFT_71116 PE=4 SV=1
MM1 pKa = 7.15PQVPARR7 pKa = 11.84PPPRR11 pKa = 11.84SSVPTEE17 pKa = 3.94SVFDD21 pKa = 4.64DD22 pKa = 4.28DD23 pKa = 4.46AASAFSFSIYY33 pKa = 10.44EE34 pKa = 3.9VDD36 pKa = 4.7LGDD39 pKa = 3.88ARR41 pKa = 11.84SGGSSSSASAYY52 pKa = 8.82SQPSFDD58 pKa = 4.38AFAAHH63 pKa = 7.21DD64 pKa = 3.76VAFDD68 pKa = 4.06LDD70 pKa = 4.1GGMMLPVSLPATPNDD85 pKa = 3.34IDD87 pKa = 6.24DD88 pKa = 3.8LTGWDD93 pKa = 4.29DD94 pKa = 3.51EE95 pKa = 5.02VEE97 pKa = 4.36FVFDD101 pKa = 3.94EE102 pKa = 4.35EE103 pKa = 4.64EE104 pKa = 4.74EE105 pKa = 4.08IHH107 pKa = 8.03SMDD110 pKa = 3.49LEE112 pKa = 4.32IYY114 pKa = 10.07IVRR117 pKa = 11.84TTINLMNSAGIYY129 pKa = 9.54IVRR132 pKa = 11.84TTRR135 pKa = 11.84DD136 pKa = 3.53DD137 pKa = 3.77YY138 pKa = 11.49YY139 pKa = 9.96EE140 pKa = 4.46TYY142 pKa = 10.62SSYY145 pKa = 10.75IYY147 pKa = 9.32RR148 pKa = 11.84TQVQQ152 pKa = 3.14
MM1 pKa = 7.15PQVPARR7 pKa = 11.84PPPRR11 pKa = 11.84SSVPTEE17 pKa = 3.94SVFDD21 pKa = 4.64DD22 pKa = 4.28DD23 pKa = 4.46AASAFSFSIYY33 pKa = 10.44EE34 pKa = 3.9VDD36 pKa = 4.7LGDD39 pKa = 3.88ARR41 pKa = 11.84SGGSSSSASAYY52 pKa = 8.82SQPSFDD58 pKa = 4.38AFAAHH63 pKa = 7.21DD64 pKa = 3.76VAFDD68 pKa = 4.06LDD70 pKa = 4.1GGMMLPVSLPATPNDD85 pKa = 3.34IDD87 pKa = 6.24DD88 pKa = 3.8LTGWDD93 pKa = 4.29DD94 pKa = 3.51EE95 pKa = 5.02VEE97 pKa = 4.36FVFDD101 pKa = 3.94EE102 pKa = 4.35EE103 pKa = 4.64EE104 pKa = 4.74EE105 pKa = 4.08IHH107 pKa = 8.03SMDD110 pKa = 3.49LEE112 pKa = 4.32IYY114 pKa = 10.07IVRR117 pKa = 11.84TTINLMNSAGIYY129 pKa = 9.54IVRR132 pKa = 11.84TTRR135 pKa = 11.84DD136 pKa = 3.53DD137 pKa = 3.77YY138 pKa = 11.49YY139 pKa = 9.96EE140 pKa = 4.46TYY142 pKa = 10.62SSYY145 pKa = 10.75IYY147 pKa = 9.32RR148 pKa = 11.84TQVQQ152 pKa = 3.14
Molecular weight: 16.85 kDa
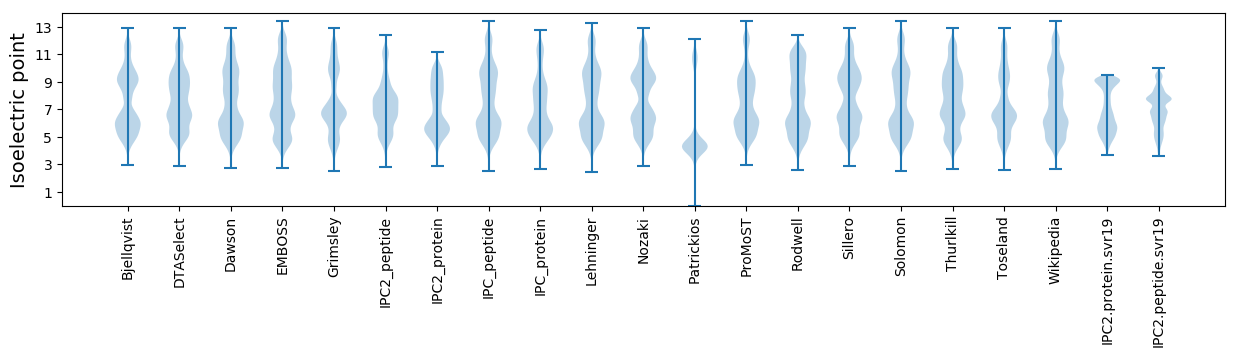
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D2PAU8|A0A0D2PAU8_HYPSF Uncharacterized protein OS=Hypholoma sublateritium (strain FD-334 SS-4) OX=945553 GN=HYPSUDRAFT_34322 PE=4 SV=1
MM1 pKa = 7.45AVSTPQTPLCHH12 pKa = 6.88ALLTTHH18 pKa = 6.83SARR21 pKa = 11.84TAPARR26 pKa = 11.84ASAATSRR33 pKa = 11.84ARR35 pKa = 11.84ASAASSTASPTRR47 pKa = 11.84ASTASARR54 pKa = 11.84SARR57 pKa = 11.84RR58 pKa = 11.84ASSAGRR64 pKa = 11.84TAARR68 pKa = 11.84TRR70 pKa = 11.84ATPPPPLPTPPSRR83 pKa = 11.84RR84 pKa = 11.84PPSPSGAQVHH94 pKa = 5.05RR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84TRR102 pKa = 11.84PAPRR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84PPPPPPRR116 pKa = 11.84CTPSRR121 pKa = 11.84STRR124 pKa = 11.84TRR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84GTTPCTTPRR138 pKa = 11.84PARR141 pKa = 11.84SCRR144 pKa = 11.84TRR146 pKa = 11.84TPRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84ARR153 pKa = 11.84APRR156 pKa = 11.84ATRR159 pKa = 11.84PRR161 pKa = 11.84LPMRR165 pKa = 11.84PRR167 pKa = 11.84TRR169 pKa = 11.84RR170 pKa = 11.84TGSRR174 pKa = 11.84SCRR177 pKa = 11.84TTSTPGGTRR186 pKa = 11.84RR187 pKa = 11.84SRR189 pKa = 11.84TTRR192 pKa = 11.84RR193 pKa = 11.84CTPRR197 pKa = 11.84SNSRR201 pKa = 11.84RR202 pKa = 11.84GRR204 pKa = 11.84RR205 pKa = 11.84PQARR209 pKa = 11.84RR210 pKa = 11.84PPPPLRR216 pKa = 11.84LHH218 pKa = 5.71PQRR221 pKa = 11.84LRR223 pKa = 11.84RR224 pKa = 11.84TRR226 pKa = 11.84PRR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84RR231 pKa = 11.84SAPLIPQAHH240 pKa = 5.48RR241 pKa = 11.84TPRR244 pKa = 11.84RR245 pKa = 11.84RR246 pKa = 11.84ARR248 pKa = 11.84SRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84PLTEE259 pKa = 3.54MGRR262 pKa = 11.84SARR265 pKa = 11.84AWARR269 pKa = 11.84TASRR273 pKa = 11.84ARR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84RR279 pKa = 11.84ARR281 pKa = 11.84RR282 pKa = 11.84ARR284 pKa = 11.84GGVRR288 pKa = 11.84RR289 pKa = 11.84RR290 pKa = 11.84MHH292 pKa = 6.3SLSHH296 pKa = 6.9PPLPSPLPLPIRR308 pKa = 11.84RR309 pKa = 11.84RR310 pKa = 11.84MAARR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84GEE318 pKa = 3.85QEE320 pKa = 3.6EE321 pKa = 4.82GG322 pKa = 3.27
MM1 pKa = 7.45AVSTPQTPLCHH12 pKa = 6.88ALLTTHH18 pKa = 6.83SARR21 pKa = 11.84TAPARR26 pKa = 11.84ASAATSRR33 pKa = 11.84ARR35 pKa = 11.84ASAASSTASPTRR47 pKa = 11.84ASTASARR54 pKa = 11.84SARR57 pKa = 11.84RR58 pKa = 11.84ASSAGRR64 pKa = 11.84TAARR68 pKa = 11.84TRR70 pKa = 11.84ATPPPPLPTPPSRR83 pKa = 11.84RR84 pKa = 11.84PPSPSGAQVHH94 pKa = 5.05RR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84RR100 pKa = 11.84TRR102 pKa = 11.84PAPRR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84PPPPPPRR116 pKa = 11.84CTPSRR121 pKa = 11.84STRR124 pKa = 11.84TRR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84GTTPCTTPRR138 pKa = 11.84PARR141 pKa = 11.84SCRR144 pKa = 11.84TRR146 pKa = 11.84TPRR149 pKa = 11.84RR150 pKa = 11.84RR151 pKa = 11.84ARR153 pKa = 11.84APRR156 pKa = 11.84ATRR159 pKa = 11.84PRR161 pKa = 11.84LPMRR165 pKa = 11.84PRR167 pKa = 11.84TRR169 pKa = 11.84RR170 pKa = 11.84TGSRR174 pKa = 11.84SCRR177 pKa = 11.84TTSTPGGTRR186 pKa = 11.84RR187 pKa = 11.84SRR189 pKa = 11.84TTRR192 pKa = 11.84RR193 pKa = 11.84CTPRR197 pKa = 11.84SNSRR201 pKa = 11.84RR202 pKa = 11.84GRR204 pKa = 11.84RR205 pKa = 11.84PQARR209 pKa = 11.84RR210 pKa = 11.84PPPPLRR216 pKa = 11.84LHH218 pKa = 5.71PQRR221 pKa = 11.84LRR223 pKa = 11.84RR224 pKa = 11.84TRR226 pKa = 11.84PRR228 pKa = 11.84RR229 pKa = 11.84RR230 pKa = 11.84RR231 pKa = 11.84SAPLIPQAHH240 pKa = 5.48RR241 pKa = 11.84TPRR244 pKa = 11.84RR245 pKa = 11.84RR246 pKa = 11.84ARR248 pKa = 11.84SRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84PLTEE259 pKa = 3.54MGRR262 pKa = 11.84SARR265 pKa = 11.84AWARR269 pKa = 11.84TASRR273 pKa = 11.84ARR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84RR278 pKa = 11.84RR279 pKa = 11.84ARR281 pKa = 11.84RR282 pKa = 11.84ARR284 pKa = 11.84GGVRR288 pKa = 11.84RR289 pKa = 11.84RR290 pKa = 11.84MHH292 pKa = 6.3SLSHH296 pKa = 6.9PPLPSPLPLPIRR308 pKa = 11.84RR309 pKa = 11.84RR310 pKa = 11.84MAARR314 pKa = 11.84RR315 pKa = 11.84RR316 pKa = 11.84GEE318 pKa = 3.85QEE320 pKa = 3.6EE321 pKa = 4.82GG322 pKa = 3.27
Molecular weight: 36.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6809349 |
49 |
3938 |
386.1 |
42.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.328 ± 0.026 | 1.369 ± 0.008 |
5.38 ± 0.015 | 5.426 ± 0.02 |
3.742 ± 0.011 | 6.34 ± 0.019 |
2.688 ± 0.01 | 5.033 ± 0.015 |
4.272 ± 0.016 | 9.122 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.024 ± 0.007 | 3.485 ± 0.01 |
6.767 ± 0.023 | 3.647 ± 0.014 |
6.458 ± 0.017 | 8.512 ± 0.025 |
6.152 ± 0.014 | 6.186 ± 0.015 |
1.387 ± 0.006 | 2.662 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
