
Streptomyces longwoodensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
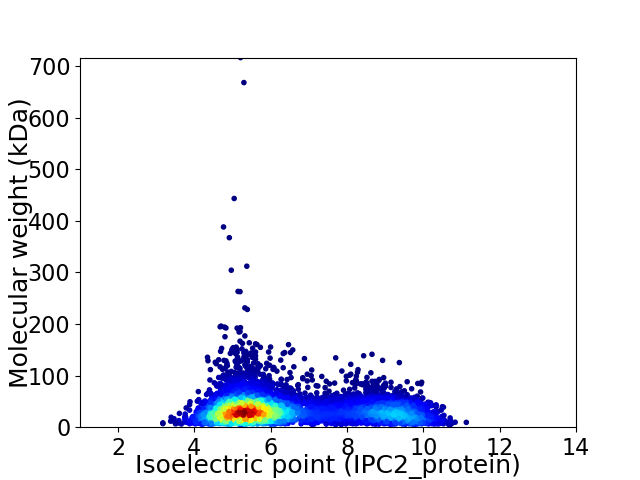
Virtual 2D-PAGE plot for 6937 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
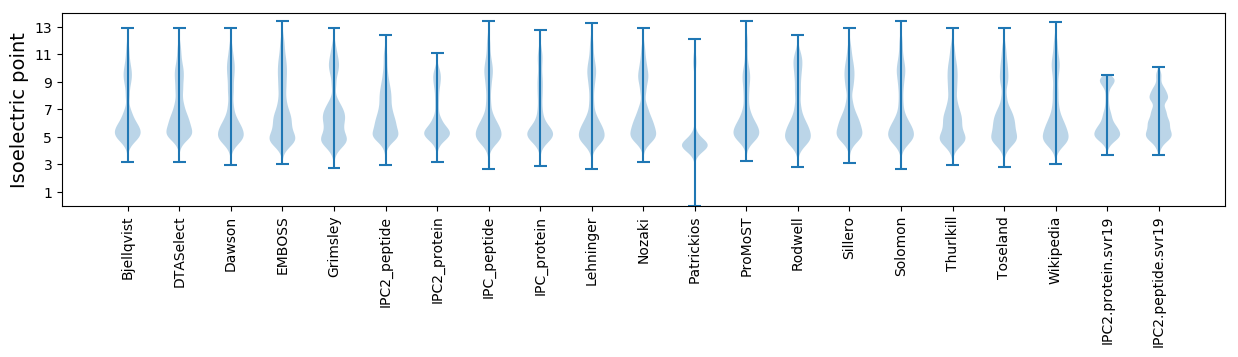
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A101QNX5|A0A101QNX5_9ACTN SnoaL-like domain-containing protein OS=Streptomyces longwoodensis OX=68231 GN=AQJ30_33935 PE=4 SV=1
MM1 pKa = 7.6SSGASGGPAARR12 pKa = 11.84YY13 pKa = 9.05LYY15 pKa = 9.73QLNGAIPLSVANAGAGTGIDD35 pKa = 3.63VTPDD39 pKa = 2.78SGTNTLTVTAVAADD53 pKa = 3.93GAIGGSTNCHH63 pKa = 6.24FFAGPPPTAPDD74 pKa = 3.69GDD76 pKa = 4.05LTGDD80 pKa = 4.17GLPDD84 pKa = 3.5LTVVGAQAGLPSGLWLARR102 pKa = 11.84GTADD106 pKa = 3.52GQVDD110 pKa = 3.84PDD112 pKa = 3.67VTNVGGQGTGASSAGSASDD131 pKa = 3.17WDD133 pKa = 3.95GTQTITGHH141 pKa = 5.57FHH143 pKa = 6.16TGAGFNDD150 pKa = 3.5VLVYY154 pKa = 10.85APTRR158 pKa = 11.84GTGTILYY165 pKa = 7.45GTGDD169 pKa = 3.9GSALSPYY176 pKa = 9.98SGHH179 pKa = 5.82EE180 pKa = 4.2VNVSSLAFTDD190 pKa = 3.76EE191 pKa = 4.38SGVTGAPGSRR201 pKa = 11.84ATSIASGGEE210 pKa = 3.88LYY212 pKa = 9.31RR213 pKa = 11.84TVNDD217 pKa = 3.49MPRR220 pKa = 11.84AGLPDD225 pKa = 3.95LLMIVNGQLWDD236 pKa = 3.91EE237 pKa = 4.85PGLMAPGAFLGLDD250 pKa = 3.09NALPLTSTNPTGSGDD265 pKa = 3.11WTGWSLTSSLVDD277 pKa = 3.23GLPALFARR285 pKa = 11.84NAATGALYY293 pKa = 10.54YY294 pKa = 7.04YY295 pKa = 9.81TPQQLQDD302 pKa = 3.73LAYY305 pKa = 9.54GNAVTPLKK313 pKa = 10.61LADD316 pKa = 3.77SGYY319 pKa = 10.92DD320 pKa = 3.23AAAVPVLQAADD331 pKa = 4.26LNSDD335 pKa = 3.81GTPDD339 pKa = 3.79LRR341 pKa = 11.84TVSATGVTTAWILDD355 pKa = 3.89PAAGTLTPRR364 pKa = 11.84PAQSLNVPGGTAA376 pKa = 3.09
MM1 pKa = 7.6SSGASGGPAARR12 pKa = 11.84YY13 pKa = 9.05LYY15 pKa = 9.73QLNGAIPLSVANAGAGTGIDD35 pKa = 3.63VTPDD39 pKa = 2.78SGTNTLTVTAVAADD53 pKa = 3.93GAIGGSTNCHH63 pKa = 6.24FFAGPPPTAPDD74 pKa = 3.69GDD76 pKa = 4.05LTGDD80 pKa = 4.17GLPDD84 pKa = 3.5LTVVGAQAGLPSGLWLARR102 pKa = 11.84GTADD106 pKa = 3.52GQVDD110 pKa = 3.84PDD112 pKa = 3.67VTNVGGQGTGASSAGSASDD131 pKa = 3.17WDD133 pKa = 3.95GTQTITGHH141 pKa = 5.57FHH143 pKa = 6.16TGAGFNDD150 pKa = 3.5VLVYY154 pKa = 10.85APTRR158 pKa = 11.84GTGTILYY165 pKa = 7.45GTGDD169 pKa = 3.9GSALSPYY176 pKa = 9.98SGHH179 pKa = 5.82EE180 pKa = 4.2VNVSSLAFTDD190 pKa = 3.76EE191 pKa = 4.38SGVTGAPGSRR201 pKa = 11.84ATSIASGGEE210 pKa = 3.88LYY212 pKa = 9.31RR213 pKa = 11.84TVNDD217 pKa = 3.49MPRR220 pKa = 11.84AGLPDD225 pKa = 3.95LLMIVNGQLWDD236 pKa = 3.91EE237 pKa = 4.85PGLMAPGAFLGLDD250 pKa = 3.09NALPLTSTNPTGSGDD265 pKa = 3.11WTGWSLTSSLVDD277 pKa = 3.23GLPALFARR285 pKa = 11.84NAATGALYY293 pKa = 10.54YY294 pKa = 7.04YY295 pKa = 9.81TPQQLQDD302 pKa = 3.73LAYY305 pKa = 9.54GNAVTPLKK313 pKa = 10.61LADD316 pKa = 3.77SGYY319 pKa = 10.92DD320 pKa = 3.23AAAVPVLQAADD331 pKa = 4.26LNSDD335 pKa = 3.81GTPDD339 pKa = 3.79LRR341 pKa = 11.84TVSATGVTTAWILDD355 pKa = 3.89PAAGTLTPRR364 pKa = 11.84PAQSLNVPGGTAA376 pKa = 3.09
Molecular weight: 37.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A117QK81|A0A117QK81_9ACTN Uncharacterized protein OS=Streptomyces longwoodensis OX=68231 GN=AQJ30_36140 PE=4 SV=1
MM1 pKa = 7.47ANCRR5 pKa = 11.84RR6 pKa = 11.84TPVTAIAPKK15 pKa = 9.93AVAPRR20 pKa = 11.84VVAMARR26 pKa = 11.84TGAVGVAVPARR37 pKa = 11.84VAVVVTRR44 pKa = 11.84PRR46 pKa = 11.84TVVSAVGRR54 pKa = 11.84VAIRR58 pKa = 11.84ALARR62 pKa = 11.84AVAMTARR69 pKa = 11.84VGARR73 pKa = 11.84RR74 pKa = 11.84LMARR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84PGAVLVGRR88 pKa = 11.84AARR91 pKa = 3.8
MM1 pKa = 7.47ANCRR5 pKa = 11.84RR6 pKa = 11.84TPVTAIAPKK15 pKa = 9.93AVAPRR20 pKa = 11.84VVAMARR26 pKa = 11.84TGAVGVAVPARR37 pKa = 11.84VAVVVTRR44 pKa = 11.84PRR46 pKa = 11.84TVVSAVGRR54 pKa = 11.84VAIRR58 pKa = 11.84ALARR62 pKa = 11.84AVAMTARR69 pKa = 11.84VGARR73 pKa = 11.84RR74 pKa = 11.84LMARR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84PGAVLVGRR88 pKa = 11.84AARR91 pKa = 3.8
Molecular weight: 9.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2309017 |
29 |
6656 |
332.9 |
35.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.955 ± 0.042 | 0.783 ± 0.008 |
6.175 ± 0.026 | 5.491 ± 0.03 |
2.629 ± 0.016 | 9.501 ± 0.032 |
2.414 ± 0.015 | 2.724 ± 0.022 |
1.932 ± 0.026 | 10.426 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.579 ± 0.012 | 1.625 ± 0.016 |
6.256 ± 0.031 | 2.773 ± 0.019 |
8.311 ± 0.034 | 4.848 ± 0.021 |
6.287 ± 0.028 | 8.692 ± 0.027 |
1.528 ± 0.014 | 2.071 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
