
Bean necrotic mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Tospoviridae; Orthotospovirus; Bean necrotic mosaic orthotospovirus
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
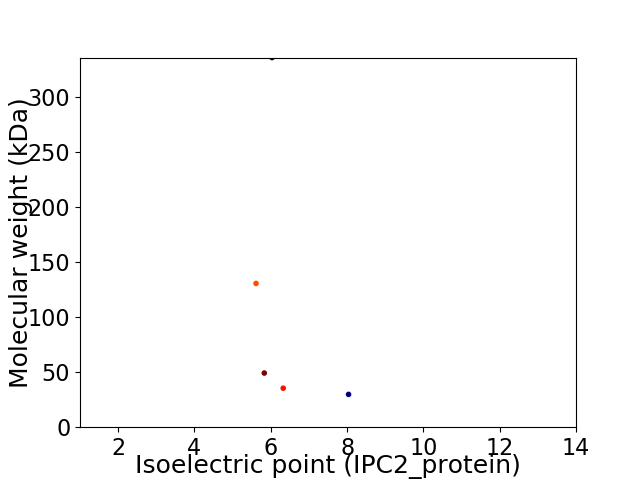
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3PCT2|I3PCT2_9VIRU Non-structural movement protein OS=Bean necrotic mosaic virus OX=1033976 GN=NSm PE=4 SV=1
MM1 pKa = 7.61GFRR4 pKa = 11.84EE5 pKa = 4.3VFVIICLFCLNLVFLDD21 pKa = 4.47FSINAHH27 pKa = 4.93TRR29 pKa = 11.84GDD31 pKa = 3.76HH32 pKa = 6.28EE33 pKa = 4.68LSQEE37 pKa = 3.74ISFEE41 pKa = 4.25DD42 pKa = 3.88LSKK45 pKa = 11.29DD46 pKa = 3.39AVEE49 pKa = 4.34SATLLGVDD57 pKa = 4.74GKK59 pKa = 10.88FKK61 pKa = 10.88SGPLLDD67 pKa = 5.34KK68 pKa = 10.9PDD70 pKa = 4.83IDD72 pKa = 5.03DD73 pKa = 3.78QMIPNRR79 pKa = 11.84GKK81 pKa = 8.94RR82 pKa = 11.84TEE84 pKa = 4.39PSSSSPPSSEE94 pKa = 3.6QLTCKK99 pKa = 10.45RR100 pKa = 11.84INSNNCKK107 pKa = 9.82IVGKK111 pKa = 8.87TSFNFYY117 pKa = 10.53YY118 pKa = 10.32QISDD122 pKa = 3.55NEE124 pKa = 4.55TTVSCLSSEE133 pKa = 4.18SQDD136 pKa = 3.53YY137 pKa = 10.71KK138 pKa = 11.16SCNPDD143 pKa = 2.72AMQEE147 pKa = 4.06FLSADD152 pKa = 3.6VLPITTMLNKK162 pKa = 9.98RR163 pKa = 11.84VLSVGSKK170 pKa = 10.26YY171 pKa = 10.66FILNSADD178 pKa = 3.32PSSYY182 pKa = 9.98PVQDD186 pKa = 3.54HH187 pKa = 6.69GSVQLGLGNIYY198 pKa = 10.21NVRR201 pKa = 11.84LTGGCNITKK210 pKa = 10.05VSLANPFYY218 pKa = 10.01ITLSSTAKK226 pKa = 10.34KK227 pKa = 10.32CLCCKK232 pKa = 10.41KK233 pKa = 10.31IGSEE237 pKa = 4.23EE238 pKa = 4.05VNLMKK243 pKa = 10.59FSSEE247 pKa = 3.75KK248 pKa = 10.19TIEE251 pKa = 4.0LSEE254 pKa = 4.74DD255 pKa = 3.71SLDD258 pKa = 3.76GTHH261 pKa = 6.92VLLCGDD267 pKa = 4.22RR268 pKa = 11.84VSEE271 pKa = 4.56IPKK274 pKa = 9.62IDD276 pKa = 3.3INKK279 pKa = 9.51RR280 pKa = 11.84NCLVKK285 pKa = 10.93YY286 pKa = 9.93KK287 pKa = 10.54GKK289 pKa = 10.46RR290 pKa = 11.84YY291 pKa = 8.06QQSACVHH298 pKa = 5.94FNLLRR303 pKa = 11.84YY304 pKa = 9.71LIVSIILFFPVSWFLNKK321 pKa = 9.54TKK323 pKa = 10.81EE324 pKa = 5.11SMFLWYY330 pKa = 9.95DD331 pKa = 2.88ILGIVLYY338 pKa = 9.88PLLYY342 pKa = 10.06ALNWIWQYY350 pKa = 11.16FPLKK354 pKa = 10.32CSCCGNFSLITHH366 pKa = 6.83KK367 pKa = 10.76CYY369 pKa = 10.44EE370 pKa = 4.13KK371 pKa = 10.77CVCNLSKK378 pKa = 10.75AKK380 pKa = 10.17KK381 pKa = 8.88EE382 pKa = 3.96HH383 pKa = 7.27AEE385 pKa = 3.98TCQILTGKK393 pKa = 8.55IFKK396 pKa = 9.59MKK398 pKa = 9.57EE399 pKa = 3.49VKK401 pKa = 10.27AEE403 pKa = 3.8KK404 pKa = 10.35RR405 pKa = 11.84EE406 pKa = 4.31AITPDD411 pKa = 3.72EE412 pKa = 4.13IVSLNSEE419 pKa = 4.09QEE421 pKa = 3.94KK422 pKa = 10.51PPVIKK427 pKa = 10.41SSRR430 pKa = 11.84FQTFQTIINTKK441 pKa = 9.71ISSEE445 pKa = 4.11TLLLITKK452 pKa = 9.77ILLGFLIFSQIPKK465 pKa = 8.83TMAMSNEE472 pKa = 3.56QALCVNYY479 pKa = 10.1CRR481 pKa = 11.84AQIGCSKK488 pKa = 10.09FLWKK492 pKa = 10.72NEE494 pKa = 3.85EE495 pKa = 4.09TCISRR500 pKa = 11.84KK501 pKa = 8.59DD502 pKa = 3.65LKK504 pKa = 10.7CNCVIGKK511 pKa = 8.41EE512 pKa = 4.25LISEE516 pKa = 3.97EE517 pKa = 4.33AYY519 pKa = 10.12INGTRR524 pKa = 11.84YY525 pKa = 9.55AVSMPNTCMDD535 pKa = 3.84DD536 pKa = 3.58TCDD539 pKa = 3.49VAEE542 pKa = 4.76GDD544 pKa = 3.87VEE546 pKa = 4.13NLIVCRR552 pKa = 11.84LGCKK556 pKa = 9.8KK557 pKa = 10.28LRR559 pKa = 11.84SLKK562 pKa = 8.65TAKK565 pKa = 9.98LSKK568 pKa = 9.48RR569 pKa = 11.84TFSTGIFSQSLSMNQIYY586 pKa = 8.05STLRR590 pKa = 11.84MQDD593 pKa = 3.19GYY595 pKa = 10.71IDD597 pKa = 3.81SLEE600 pKa = 4.22ALSTLGDD607 pKa = 3.77PPSDD611 pKa = 3.07KK612 pKa = 10.71VVQIDD617 pKa = 3.59DD618 pKa = 5.51DD619 pKa = 4.02IPEE622 pKa = 5.29GILPRR627 pKa = 11.84QSFVYY632 pKa = 10.48SSVVDD637 pKa = 3.84GKK639 pKa = 10.96YY640 pKa = 9.88RR641 pKa = 11.84YY642 pKa = 10.12LMSFDD647 pKa = 5.65AIQSSGYY654 pKa = 8.56VYY656 pKa = 10.95SMKK659 pKa = 9.71MLKK662 pKa = 8.59WHH664 pKa = 7.13PPQEE668 pKa = 3.85LVVYY672 pKa = 9.66VKK674 pKa = 10.52EE675 pKa = 4.22AGVTYY680 pKa = 9.21TIKK683 pKa = 10.48PLYY686 pKa = 8.21HH687 pKa = 6.44TAPISSTHH695 pKa = 3.84THH697 pKa = 6.18VYY699 pKa = 6.48TTCTGNCEE707 pKa = 4.41TCRR710 pKa = 11.84KK711 pKa = 7.53EE712 pKa = 4.37HH713 pKa = 6.76PLSGYY718 pKa = 9.12QDD720 pKa = 3.78YY721 pKa = 11.11CISPTSNWGCEE732 pKa = 4.03EE733 pKa = 4.63LGCLAIGEE741 pKa = 4.66GSTCGYY747 pKa = 9.08CRR749 pKa = 11.84NVYY752 pKa = 9.95DD753 pKa = 4.19LSKK756 pKa = 9.92MYY758 pKa = 10.51SINQVLSSHH767 pKa = 5.38VTVKK771 pKa = 10.58VCFKK775 pKa = 10.39GFAGATCQTITDD787 pKa = 4.85EE788 pKa = 4.58IPYY791 pKa = 9.79QNSYY795 pKa = 9.12YY796 pKa = 10.34QISLDD801 pKa = 3.9ADD803 pKa = 3.8LHH805 pKa = 6.72NDD807 pKa = 3.56DD808 pKa = 5.42LGSGSRR814 pKa = 11.84IAIDD818 pKa = 3.64PNGLIMKK825 pKa = 9.71GNIANLKK832 pKa = 10.19DD833 pKa = 3.1SSTAFGHH840 pKa = 5.9PQLDD844 pKa = 3.63KK845 pKa = 10.87TGKK848 pKa = 10.32LLFGKK853 pKa = 10.51ANLDD857 pKa = 3.78LNDD860 pKa = 4.07FTWSCAVIGSKK871 pKa = 10.46HH872 pKa = 5.9VDD874 pKa = 3.02IKK876 pKa = 11.06KK877 pKa = 10.21CGYY880 pKa = 8.35DD881 pKa = 3.18TYY883 pKa = 10.7HH884 pKa = 6.8QYY886 pKa = 10.67IGLEE890 pKa = 4.46PISNHH895 pKa = 5.13YY896 pKa = 10.4ASEE899 pKa = 4.08ISDD902 pKa = 3.17EE903 pKa = 4.13KK904 pKa = 11.45LFVKK908 pKa = 10.41KK909 pKa = 10.34DD910 pKa = 3.81FKK912 pKa = 11.01VGKK915 pKa = 9.92INLIVDD921 pKa = 4.46LPSEE925 pKa = 4.35LFKK928 pKa = 11.3QPAKK932 pKa = 10.09KK933 pKa = 10.15PKK935 pKa = 9.4VSLINSEE942 pKa = 4.45CKK944 pKa = 10.48GCLSCGSGIKK954 pKa = 9.1CTLEE958 pKa = 3.87MTSDD962 pKa = 3.91SIFAGNINWEE972 pKa = 4.33HH973 pKa = 6.19CTSEE977 pKa = 4.11PEE979 pKa = 4.05YY980 pKa = 10.69IAMKK984 pKa = 10.11KK985 pKa = 9.89GSNIIKK991 pKa = 10.53VNMFCLKK998 pKa = 10.5NPSLTKK1004 pKa = 10.1MILEE1008 pKa = 4.27PSGDD1012 pKa = 3.52SDD1014 pKa = 4.67LSLEE1018 pKa = 4.14FEE1020 pKa = 4.31TRR1022 pKa = 11.84NVEE1025 pKa = 4.14IIDD1028 pKa = 3.82PEE1030 pKa = 4.43TIIDD1034 pKa = 3.98HH1035 pKa = 6.9NDD1037 pKa = 3.07DD1038 pKa = 3.8AFNEE1042 pKa = 4.22EE1043 pKa = 3.5VDD1045 pKa = 3.8YY1046 pKa = 11.77DD1047 pKa = 3.76NDD1049 pKa = 3.75TSFKK1053 pKa = 11.12SLWDD1057 pKa = 3.63YY1058 pKa = 11.37LKK1060 pKa = 11.28SPFNWIASFFGDD1072 pKa = 3.47FFEE1075 pKa = 4.37IVRR1078 pKa = 11.84VLLVLACCVFAAICLSKK1095 pKa = 10.74LFEE1098 pKa = 4.19ICRR1101 pKa = 11.84EE1102 pKa = 4.08YY1103 pKa = 11.12YY1104 pKa = 9.97KK1105 pKa = 10.29QEE1107 pKa = 3.98HH1108 pKa = 5.84YY1109 pKa = 10.39KK1110 pKa = 10.79KK1111 pKa = 10.41SVKK1114 pKa = 10.14SDD1116 pKa = 3.32LKK1118 pKa = 11.59DD1119 pKa = 3.23MDD1121 pKa = 4.4EE1122 pKa = 4.95EE1123 pKa = 5.15DD1124 pKa = 4.51PEE1126 pKa = 4.79DD1127 pKa = 4.63PEE1129 pKa = 7.43DD1130 pKa = 4.32ILGNLKK1136 pKa = 9.95NQITEE1141 pKa = 3.87DD1142 pKa = 3.03TLYY1145 pKa = 10.88RR1146 pKa = 11.84RR1147 pKa = 11.84NLKK1150 pKa = 9.62PKK1152 pKa = 9.93KK1153 pKa = 10.3SPFDD1157 pKa = 3.52SVMTT1161 pKa = 4.39
MM1 pKa = 7.61GFRR4 pKa = 11.84EE5 pKa = 4.3VFVIICLFCLNLVFLDD21 pKa = 4.47FSINAHH27 pKa = 4.93TRR29 pKa = 11.84GDD31 pKa = 3.76HH32 pKa = 6.28EE33 pKa = 4.68LSQEE37 pKa = 3.74ISFEE41 pKa = 4.25DD42 pKa = 3.88LSKK45 pKa = 11.29DD46 pKa = 3.39AVEE49 pKa = 4.34SATLLGVDD57 pKa = 4.74GKK59 pKa = 10.88FKK61 pKa = 10.88SGPLLDD67 pKa = 5.34KK68 pKa = 10.9PDD70 pKa = 4.83IDD72 pKa = 5.03DD73 pKa = 3.78QMIPNRR79 pKa = 11.84GKK81 pKa = 8.94RR82 pKa = 11.84TEE84 pKa = 4.39PSSSSPPSSEE94 pKa = 3.6QLTCKK99 pKa = 10.45RR100 pKa = 11.84INSNNCKK107 pKa = 9.82IVGKK111 pKa = 8.87TSFNFYY117 pKa = 10.53YY118 pKa = 10.32QISDD122 pKa = 3.55NEE124 pKa = 4.55TTVSCLSSEE133 pKa = 4.18SQDD136 pKa = 3.53YY137 pKa = 10.71KK138 pKa = 11.16SCNPDD143 pKa = 2.72AMQEE147 pKa = 4.06FLSADD152 pKa = 3.6VLPITTMLNKK162 pKa = 9.98RR163 pKa = 11.84VLSVGSKK170 pKa = 10.26YY171 pKa = 10.66FILNSADD178 pKa = 3.32PSSYY182 pKa = 9.98PVQDD186 pKa = 3.54HH187 pKa = 6.69GSVQLGLGNIYY198 pKa = 10.21NVRR201 pKa = 11.84LTGGCNITKK210 pKa = 10.05VSLANPFYY218 pKa = 10.01ITLSSTAKK226 pKa = 10.34KK227 pKa = 10.32CLCCKK232 pKa = 10.41KK233 pKa = 10.31IGSEE237 pKa = 4.23EE238 pKa = 4.05VNLMKK243 pKa = 10.59FSSEE247 pKa = 3.75KK248 pKa = 10.19TIEE251 pKa = 4.0LSEE254 pKa = 4.74DD255 pKa = 3.71SLDD258 pKa = 3.76GTHH261 pKa = 6.92VLLCGDD267 pKa = 4.22RR268 pKa = 11.84VSEE271 pKa = 4.56IPKK274 pKa = 9.62IDD276 pKa = 3.3INKK279 pKa = 9.51RR280 pKa = 11.84NCLVKK285 pKa = 10.93YY286 pKa = 9.93KK287 pKa = 10.54GKK289 pKa = 10.46RR290 pKa = 11.84YY291 pKa = 8.06QQSACVHH298 pKa = 5.94FNLLRR303 pKa = 11.84YY304 pKa = 9.71LIVSIILFFPVSWFLNKK321 pKa = 9.54TKK323 pKa = 10.81EE324 pKa = 5.11SMFLWYY330 pKa = 9.95DD331 pKa = 2.88ILGIVLYY338 pKa = 9.88PLLYY342 pKa = 10.06ALNWIWQYY350 pKa = 11.16FPLKK354 pKa = 10.32CSCCGNFSLITHH366 pKa = 6.83KK367 pKa = 10.76CYY369 pKa = 10.44EE370 pKa = 4.13KK371 pKa = 10.77CVCNLSKK378 pKa = 10.75AKK380 pKa = 10.17KK381 pKa = 8.88EE382 pKa = 3.96HH383 pKa = 7.27AEE385 pKa = 3.98TCQILTGKK393 pKa = 8.55IFKK396 pKa = 9.59MKK398 pKa = 9.57EE399 pKa = 3.49VKK401 pKa = 10.27AEE403 pKa = 3.8KK404 pKa = 10.35RR405 pKa = 11.84EE406 pKa = 4.31AITPDD411 pKa = 3.72EE412 pKa = 4.13IVSLNSEE419 pKa = 4.09QEE421 pKa = 3.94KK422 pKa = 10.51PPVIKK427 pKa = 10.41SSRR430 pKa = 11.84FQTFQTIINTKK441 pKa = 9.71ISSEE445 pKa = 4.11TLLLITKK452 pKa = 9.77ILLGFLIFSQIPKK465 pKa = 8.83TMAMSNEE472 pKa = 3.56QALCVNYY479 pKa = 10.1CRR481 pKa = 11.84AQIGCSKK488 pKa = 10.09FLWKK492 pKa = 10.72NEE494 pKa = 3.85EE495 pKa = 4.09TCISRR500 pKa = 11.84KK501 pKa = 8.59DD502 pKa = 3.65LKK504 pKa = 10.7CNCVIGKK511 pKa = 8.41EE512 pKa = 4.25LISEE516 pKa = 3.97EE517 pKa = 4.33AYY519 pKa = 10.12INGTRR524 pKa = 11.84YY525 pKa = 9.55AVSMPNTCMDD535 pKa = 3.84DD536 pKa = 3.58TCDD539 pKa = 3.49VAEE542 pKa = 4.76GDD544 pKa = 3.87VEE546 pKa = 4.13NLIVCRR552 pKa = 11.84LGCKK556 pKa = 9.8KK557 pKa = 10.28LRR559 pKa = 11.84SLKK562 pKa = 8.65TAKK565 pKa = 9.98LSKK568 pKa = 9.48RR569 pKa = 11.84TFSTGIFSQSLSMNQIYY586 pKa = 8.05STLRR590 pKa = 11.84MQDD593 pKa = 3.19GYY595 pKa = 10.71IDD597 pKa = 3.81SLEE600 pKa = 4.22ALSTLGDD607 pKa = 3.77PPSDD611 pKa = 3.07KK612 pKa = 10.71VVQIDD617 pKa = 3.59DD618 pKa = 5.51DD619 pKa = 4.02IPEE622 pKa = 5.29GILPRR627 pKa = 11.84QSFVYY632 pKa = 10.48SSVVDD637 pKa = 3.84GKK639 pKa = 10.96YY640 pKa = 9.88RR641 pKa = 11.84YY642 pKa = 10.12LMSFDD647 pKa = 5.65AIQSSGYY654 pKa = 8.56VYY656 pKa = 10.95SMKK659 pKa = 9.71MLKK662 pKa = 8.59WHH664 pKa = 7.13PPQEE668 pKa = 3.85LVVYY672 pKa = 9.66VKK674 pKa = 10.52EE675 pKa = 4.22AGVTYY680 pKa = 9.21TIKK683 pKa = 10.48PLYY686 pKa = 8.21HH687 pKa = 6.44TAPISSTHH695 pKa = 3.84THH697 pKa = 6.18VYY699 pKa = 6.48TTCTGNCEE707 pKa = 4.41TCRR710 pKa = 11.84KK711 pKa = 7.53EE712 pKa = 4.37HH713 pKa = 6.76PLSGYY718 pKa = 9.12QDD720 pKa = 3.78YY721 pKa = 11.11CISPTSNWGCEE732 pKa = 4.03EE733 pKa = 4.63LGCLAIGEE741 pKa = 4.66GSTCGYY747 pKa = 9.08CRR749 pKa = 11.84NVYY752 pKa = 9.95DD753 pKa = 4.19LSKK756 pKa = 9.92MYY758 pKa = 10.51SINQVLSSHH767 pKa = 5.38VTVKK771 pKa = 10.58VCFKK775 pKa = 10.39GFAGATCQTITDD787 pKa = 4.85EE788 pKa = 4.58IPYY791 pKa = 9.79QNSYY795 pKa = 9.12YY796 pKa = 10.34QISLDD801 pKa = 3.9ADD803 pKa = 3.8LHH805 pKa = 6.72NDD807 pKa = 3.56DD808 pKa = 5.42LGSGSRR814 pKa = 11.84IAIDD818 pKa = 3.64PNGLIMKK825 pKa = 9.71GNIANLKK832 pKa = 10.19DD833 pKa = 3.1SSTAFGHH840 pKa = 5.9PQLDD844 pKa = 3.63KK845 pKa = 10.87TGKK848 pKa = 10.32LLFGKK853 pKa = 10.51ANLDD857 pKa = 3.78LNDD860 pKa = 4.07FTWSCAVIGSKK871 pKa = 10.46HH872 pKa = 5.9VDD874 pKa = 3.02IKK876 pKa = 11.06KK877 pKa = 10.21CGYY880 pKa = 8.35DD881 pKa = 3.18TYY883 pKa = 10.7HH884 pKa = 6.8QYY886 pKa = 10.67IGLEE890 pKa = 4.46PISNHH895 pKa = 5.13YY896 pKa = 10.4ASEE899 pKa = 4.08ISDD902 pKa = 3.17EE903 pKa = 4.13KK904 pKa = 11.45LFVKK908 pKa = 10.41KK909 pKa = 10.34DD910 pKa = 3.81FKK912 pKa = 11.01VGKK915 pKa = 9.92INLIVDD921 pKa = 4.46LPSEE925 pKa = 4.35LFKK928 pKa = 11.3QPAKK932 pKa = 10.09KK933 pKa = 10.15PKK935 pKa = 9.4VSLINSEE942 pKa = 4.45CKK944 pKa = 10.48GCLSCGSGIKK954 pKa = 9.1CTLEE958 pKa = 3.87MTSDD962 pKa = 3.91SIFAGNINWEE972 pKa = 4.33HH973 pKa = 6.19CTSEE977 pKa = 4.11PEE979 pKa = 4.05YY980 pKa = 10.69IAMKK984 pKa = 10.11KK985 pKa = 9.89GSNIIKK991 pKa = 10.53VNMFCLKK998 pKa = 10.5NPSLTKK1004 pKa = 10.1MILEE1008 pKa = 4.27PSGDD1012 pKa = 3.52SDD1014 pKa = 4.67LSLEE1018 pKa = 4.14FEE1020 pKa = 4.31TRR1022 pKa = 11.84NVEE1025 pKa = 4.14IIDD1028 pKa = 3.82PEE1030 pKa = 4.43TIIDD1034 pKa = 3.98HH1035 pKa = 6.9NDD1037 pKa = 3.07DD1038 pKa = 3.8AFNEE1042 pKa = 4.22EE1043 pKa = 3.5VDD1045 pKa = 3.8YY1046 pKa = 11.77DD1047 pKa = 3.76NDD1049 pKa = 3.75TSFKK1053 pKa = 11.12SLWDD1057 pKa = 3.63YY1058 pKa = 11.37LKK1060 pKa = 11.28SPFNWIASFFGDD1072 pKa = 3.47FFEE1075 pKa = 4.37IVRR1078 pKa = 11.84VLLVLACCVFAAICLSKK1095 pKa = 10.74LFEE1098 pKa = 4.19ICRR1101 pKa = 11.84EE1102 pKa = 4.08YY1103 pKa = 11.12YY1104 pKa = 9.97KK1105 pKa = 10.29QEE1107 pKa = 3.98HH1108 pKa = 5.84YY1109 pKa = 10.39KK1110 pKa = 10.79KK1111 pKa = 10.41SVKK1114 pKa = 10.14SDD1116 pKa = 3.32LKK1118 pKa = 11.59DD1119 pKa = 3.23MDD1121 pKa = 4.4EE1122 pKa = 4.95EE1123 pKa = 5.15DD1124 pKa = 4.51PEE1126 pKa = 4.79DD1127 pKa = 4.63PEE1129 pKa = 7.43DD1130 pKa = 4.32ILGNLKK1136 pKa = 9.95NQITEE1141 pKa = 3.87DD1142 pKa = 3.03TLYY1145 pKa = 10.88RR1146 pKa = 11.84RR1147 pKa = 11.84NLKK1150 pKa = 9.62PKK1152 pKa = 9.93KK1153 pKa = 10.3SPFDD1157 pKa = 3.52SVMTT1161 pKa = 4.39
Molecular weight: 130.71 kDa
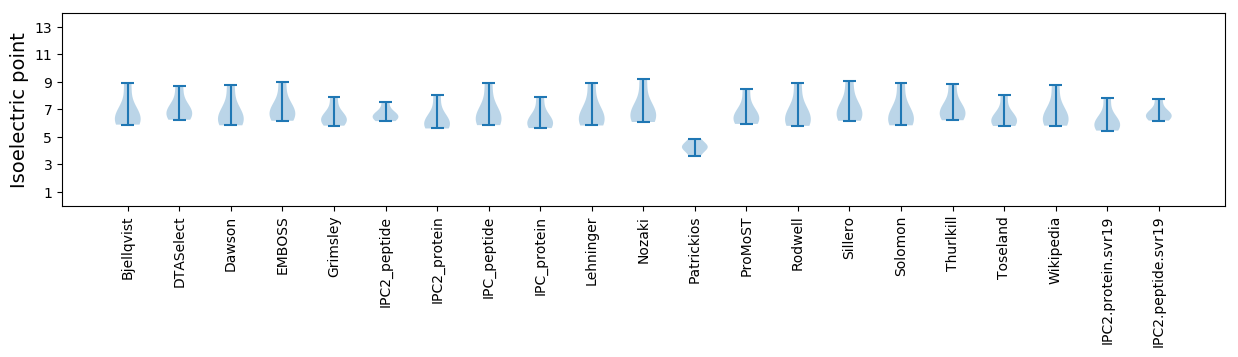
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3PCT0|I3PCT0_9VIRU Non-structural silencing protein OS=Bean necrotic mosaic virus OX=1033976 GN=NSs PE=4 SV=1
MM1 pKa = 7.62GSNALKK7 pKa = 10.91LNAEE11 pKa = 4.11NLKK14 pKa = 10.72KK15 pKa = 10.66LLLFNDD21 pKa = 3.6EE22 pKa = 4.85VEE24 pKa = 4.98FEE26 pKa = 4.17QQSTGFKK33 pKa = 10.56FVDD36 pKa = 4.12FCKK39 pKa = 10.43AHH41 pKa = 6.73EE42 pKa = 4.21KK43 pKa = 11.11DD44 pKa = 3.38KK45 pKa = 11.32FNPTSALTFLKK56 pKa = 10.23NRR58 pKa = 11.84KK59 pKa = 9.03AIYY62 pKa = 9.37SLCKK66 pKa = 10.41ASTFNYY72 pKa = 9.58GVYY75 pKa = 9.98EE76 pKa = 3.99IKK78 pKa = 10.68KK79 pKa = 9.76GDD81 pKa = 3.58KK82 pKa = 10.15ATDD85 pKa = 3.02TDD87 pKa = 4.46FTFKK91 pKa = 10.94RR92 pKa = 11.84LDD94 pKa = 2.99AFLRR98 pKa = 11.84VKK100 pKa = 10.82LLTHH104 pKa = 6.11CTKK107 pKa = 10.11IWDD110 pKa = 4.08GTDD113 pKa = 3.41PNAQTALCSEE123 pKa = 4.7LAKK126 pKa = 10.52IPIVKK131 pKa = 10.23AYY133 pKa = 9.87GLEE136 pKa = 4.08VSDD139 pKa = 4.84KK140 pKa = 8.6QKK142 pKa = 11.11CYY144 pKa = 10.27IAIVLGGNLSLLASLPGCEE163 pKa = 4.22VACFALAIFQDD174 pKa = 3.87LKK176 pKa = 11.25KK177 pKa = 10.61NEE179 pKa = 4.5LGIKK183 pKa = 10.03SDD185 pKa = 5.3FDD187 pKa = 3.91TKK189 pKa = 11.04DD190 pKa = 2.87QAGRR194 pKa = 11.84VAAVLDD200 pKa = 3.69AKK202 pKa = 10.92NFVFGEE208 pKa = 4.22PEE210 pKa = 3.69NEE212 pKa = 4.03KK213 pKa = 10.54LKK215 pKa = 10.92KK216 pKa = 9.43IAGILKK222 pKa = 10.03EE223 pKa = 4.03MTPTRR228 pKa = 11.84RR229 pKa = 11.84GHH231 pKa = 6.78AALTKK236 pKa = 10.07YY237 pKa = 10.63AEE239 pKa = 4.1QLSIISGVIGVTFEE253 pKa = 4.04MPGTSKK259 pKa = 10.94AKK261 pKa = 9.39EE262 pKa = 4.31SKK264 pKa = 10.18NHH266 pKa = 5.44GFMSS270 pKa = 4.01
MM1 pKa = 7.62GSNALKK7 pKa = 10.91LNAEE11 pKa = 4.11NLKK14 pKa = 10.72KK15 pKa = 10.66LLLFNDD21 pKa = 3.6EE22 pKa = 4.85VEE24 pKa = 4.98FEE26 pKa = 4.17QQSTGFKK33 pKa = 10.56FVDD36 pKa = 4.12FCKK39 pKa = 10.43AHH41 pKa = 6.73EE42 pKa = 4.21KK43 pKa = 11.11DD44 pKa = 3.38KK45 pKa = 11.32FNPTSALTFLKK56 pKa = 10.23NRR58 pKa = 11.84KK59 pKa = 9.03AIYY62 pKa = 9.37SLCKK66 pKa = 10.41ASTFNYY72 pKa = 9.58GVYY75 pKa = 9.98EE76 pKa = 3.99IKK78 pKa = 10.68KK79 pKa = 9.76GDD81 pKa = 3.58KK82 pKa = 10.15ATDD85 pKa = 3.02TDD87 pKa = 4.46FTFKK91 pKa = 10.94RR92 pKa = 11.84LDD94 pKa = 2.99AFLRR98 pKa = 11.84VKK100 pKa = 10.82LLTHH104 pKa = 6.11CTKK107 pKa = 10.11IWDD110 pKa = 4.08GTDD113 pKa = 3.41PNAQTALCSEE123 pKa = 4.7LAKK126 pKa = 10.52IPIVKK131 pKa = 10.23AYY133 pKa = 9.87GLEE136 pKa = 4.08VSDD139 pKa = 4.84KK140 pKa = 8.6QKK142 pKa = 11.11CYY144 pKa = 10.27IAIVLGGNLSLLASLPGCEE163 pKa = 4.22VACFALAIFQDD174 pKa = 3.87LKK176 pKa = 11.25KK177 pKa = 10.61NEE179 pKa = 4.5LGIKK183 pKa = 10.03SDD185 pKa = 5.3FDD187 pKa = 3.91TKK189 pKa = 11.04DD190 pKa = 2.87QAGRR194 pKa = 11.84VAAVLDD200 pKa = 3.69AKK202 pKa = 10.92NFVFGEE208 pKa = 4.22PEE210 pKa = 3.69NEE212 pKa = 4.03KK213 pKa = 10.54LKK215 pKa = 10.92KK216 pKa = 9.43IAGILKK222 pKa = 10.03EE223 pKa = 4.03MTPTRR228 pKa = 11.84RR229 pKa = 11.84GHH231 pKa = 6.78AALTKK236 pKa = 10.07YY237 pKa = 10.63AEE239 pKa = 4.1QLSIISGVIGVTFEE253 pKa = 4.04MPGTSKK259 pKa = 10.94AKK261 pKa = 9.39EE262 pKa = 4.31SKK264 pKa = 10.18NHH266 pKa = 5.44GFMSS270 pKa = 4.01
Molecular weight: 29.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5119 |
270 |
2932 |
1023.8 |
116.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.77 ± 0.726 | 2.559 ± 0.664 |
5.958 ± 0.247 | 6.896 ± 0.354 |
4.61 ± 0.415 | 4.884 ± 0.614 |
1.739 ± 0.15 | 7.111 ± 0.249 |
9.142 ± 0.449 | 9.807 ± 0.411 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.087 ± 0.554 | 5.997 ± 0.379 |
3.028 ± 0.38 | 2.774 ± 0.126 |
3.126 ± 0.431 | 9.631 ± 0.544 |
5.802 ± 0.417 | 5.45 ± 0.532 |
0.664 ± 0.165 | 3.966 ± 0.369 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
