
Podarcis muralis (Wall lizard) (Lacerta muralis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria; Lepi
Average proteome isoelectric point is 6.77
Get precalculated fractions of proteins
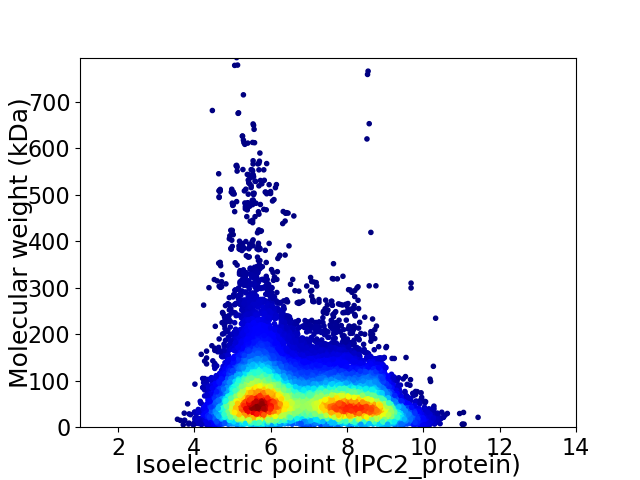
Virtual 2D-PAGE plot for 36445 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A670JAM0|A0A670JAM0_PODMU Reverse transcriptase domain-containing protein OS=Podarcis muralis OX=64176 PE=4 SV=1
MM1 pKa = 7.5WPCPLLCCLSPLLFLCLLFLPSGHH25 pKa = 7.2LCPSVCSCMDD35 pKa = 3.26YY36 pKa = 8.82HH37 pKa = 7.22TIDD40 pKa = 4.7CRR42 pKa = 11.84DD43 pKa = 3.12QGLPRR48 pKa = 11.84VPSPFPLDD56 pKa = 3.48VRR58 pKa = 11.84KK59 pKa = 9.76LLIADD64 pKa = 3.87NNIQEE69 pKa = 4.19IPGDD73 pKa = 3.61FFIFYY78 pKa = 10.58SDD80 pKa = 4.06LVYY83 pKa = 11.04LDD85 pKa = 4.07FRR87 pKa = 11.84NNSIATLQDD96 pKa = 3.16GTFSSSSKK104 pKa = 10.51LVYY107 pKa = 10.55LDD109 pKa = 4.63LSYY112 pKa = 11.96NNLTQLDD119 pKa = 3.58AGIFKK124 pKa = 10.62SAEE127 pKa = 3.94KK128 pKa = 10.38LIKK131 pKa = 10.55LSLGNNNLAEE141 pKa = 4.06VDD143 pKa = 3.65EE144 pKa = 4.83AAFEE148 pKa = 4.43SLKK151 pKa = 10.37QLQVLEE157 pKa = 5.01LNDD160 pKa = 4.57NNLQTLNVATFDD172 pKa = 3.74ALPNLRR178 pKa = 11.84TIRR181 pKa = 11.84LEE183 pKa = 4.19GNPWLCDD190 pKa = 3.4CDD192 pKa = 4.08FASLFSWLEE201 pKa = 3.55ANAPKK206 pKa = 10.16LQKK209 pKa = 10.87GLDD212 pKa = 4.03EE213 pKa = 4.48IQCTVPVEE221 pKa = 4.34EE222 pKa = 3.98ITIFLSEE229 pKa = 4.07LSEE232 pKa = 4.67ASFRR236 pKa = 11.84DD237 pKa = 3.87CKK239 pKa = 10.78FSLSLTDD246 pKa = 4.77LIIIIFSGVAVSIAAIVSSFVLALIVNCFQRR277 pKa = 11.84CAPSKK282 pKa = 11.06DD283 pKa = 3.52DD284 pKa = 6.59DD285 pKa = 4.48EE286 pKa = 7.61DD287 pKa = 4.99EE288 pKa = 5.98DD289 pKa = 6.53DD290 pKa = 5.86DD291 pKa = 5.34DD292 pKa = 4.57
MM1 pKa = 7.5WPCPLLCCLSPLLFLCLLFLPSGHH25 pKa = 7.2LCPSVCSCMDD35 pKa = 3.26YY36 pKa = 8.82HH37 pKa = 7.22TIDD40 pKa = 4.7CRR42 pKa = 11.84DD43 pKa = 3.12QGLPRR48 pKa = 11.84VPSPFPLDD56 pKa = 3.48VRR58 pKa = 11.84KK59 pKa = 9.76LLIADD64 pKa = 3.87NNIQEE69 pKa = 4.19IPGDD73 pKa = 3.61FFIFYY78 pKa = 10.58SDD80 pKa = 4.06LVYY83 pKa = 11.04LDD85 pKa = 4.07FRR87 pKa = 11.84NNSIATLQDD96 pKa = 3.16GTFSSSSKK104 pKa = 10.51LVYY107 pKa = 10.55LDD109 pKa = 4.63LSYY112 pKa = 11.96NNLTQLDD119 pKa = 3.58AGIFKK124 pKa = 10.62SAEE127 pKa = 3.94KK128 pKa = 10.38LIKK131 pKa = 10.55LSLGNNNLAEE141 pKa = 4.06VDD143 pKa = 3.65EE144 pKa = 4.83AAFEE148 pKa = 4.43SLKK151 pKa = 10.37QLQVLEE157 pKa = 5.01LNDD160 pKa = 4.57NNLQTLNVATFDD172 pKa = 3.74ALPNLRR178 pKa = 11.84TIRR181 pKa = 11.84LEE183 pKa = 4.19GNPWLCDD190 pKa = 3.4CDD192 pKa = 4.08FASLFSWLEE201 pKa = 3.55ANAPKK206 pKa = 10.16LQKK209 pKa = 10.87GLDD212 pKa = 4.03EE213 pKa = 4.48IQCTVPVEE221 pKa = 4.34EE222 pKa = 3.98ITIFLSEE229 pKa = 4.07LSEE232 pKa = 4.67ASFRR236 pKa = 11.84DD237 pKa = 3.87CKK239 pKa = 10.78FSLSLTDD246 pKa = 4.77LIIIIFSGVAVSIAAIVSSFVLALIVNCFQRR277 pKa = 11.84CAPSKK282 pKa = 11.06DD283 pKa = 3.52DD284 pKa = 6.59DD285 pKa = 4.48EE286 pKa = 7.61DD287 pKa = 4.99EE288 pKa = 5.98DD289 pKa = 6.53DD290 pKa = 5.86DD291 pKa = 5.34DD292 pKa = 4.57
Molecular weight: 32.53 kDa
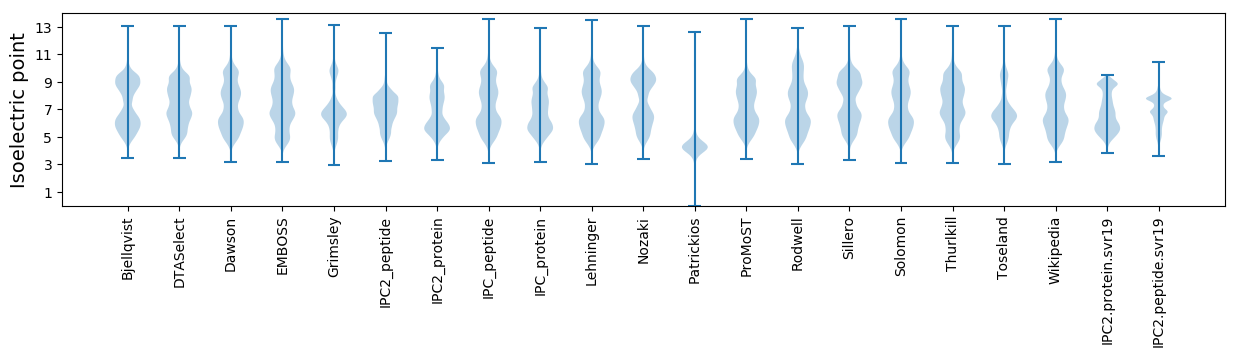
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A670KF72|A0A670KF72_PODMU KRAB domain-containing protein OS=Podarcis muralis OX=64176 PE=4 SV=1
KKK2 pKa = 9.19KKK4 pKa = 10.14RR5 pKa = 11.84VAARR9 pKa = 11.84KKK11 pKa = 7.45KKK13 pKa = 9.95RR14 pKa = 11.84VAARR18 pKa = 11.84KKK20 pKa = 7.35KKK22 pKa = 9.9RR23 pKa = 11.84VSARR27 pKa = 11.84KKK29 pKa = 8.37KKK31 pKa = 10.01RR32 pKa = 11.84VSARR36 pKa = 11.84KKK38 pKa = 8.52KKK40 pKa = 10.04RR41 pKa = 11.84VAARR45 pKa = 11.84KKK47 pKa = 7.45KKK49 pKa = 9.95RR50 pKa = 11.84VAARR54 pKa = 11.84KKK56 pKa = 7.35KKK58 pKa = 9.9RR59 pKa = 11.84VSARR63 pKa = 11.84KKK65 pKa = 8.52KKK67 pKa = 10.04RR68 pKa = 11.84VAARR72 pKa = 11.84KKK74 pKa = 7.45KKK76 pKa = 9.95RR77 pKa = 11.84VAARR81 pKa = 11.84KKK83 pKa = 7.35KKK85 pKa = 9.9RR86 pKa = 11.84VSARR90 pKa = 11.84KKK92 pKa = 8.52KKK94 pKa = 10.04RR95 pKa = 11.84VAARR99 pKa = 11.84KKK101 pKa = 7.45KKK103 pKa = 9.95RR104 pKa = 11.84VAARR108 pKa = 11.84KKK110 pKa = 7.35KKK112 pKa = 9.9RR113 pKa = 11.84VSARR117 pKa = 11.84KKK119 pKa = 8.52KKK121 pKa = 10.04RR122 pKa = 11.84VAARR126 pKa = 11.84KKK128 pKa = 7.45KKK130 pKa = 9.95RR131 pKa = 11.84VAARR135 pKa = 11.84KKK137 pKa = 7.35KKK139 pKa = 9.9RR140 pKa = 11.84VSARR144 pKa = 11.84KKK146 pKa = 8.52KKK148 pKa = 10.04RR149 pKa = 11.84VAARR153 pKa = 11.84KKK155 pKa = 7.45KKK157 pKa = 9.95RR158 pKa = 11.84VAARR162 pKa = 11.84KKK164 pKa = 7.35KKK166 pKa = 9.9RR167 pKa = 11.84VSARR171 pKa = 11.84KKK173 pKa = 8.66KKK175 pKa = 9.76QYYY178 pKa = 10.11FKKK181 pKa = 10.61NPSVGPQL
KKK2 pKa = 9.19KKK4 pKa = 10.14RR5 pKa = 11.84VAARR9 pKa = 11.84KKK11 pKa = 7.45KKK13 pKa = 9.95RR14 pKa = 11.84VAARR18 pKa = 11.84KKK20 pKa = 7.35KKK22 pKa = 9.9RR23 pKa = 11.84VSARR27 pKa = 11.84KKK29 pKa = 8.37KKK31 pKa = 10.01RR32 pKa = 11.84VSARR36 pKa = 11.84KKK38 pKa = 8.52KKK40 pKa = 10.04RR41 pKa = 11.84VAARR45 pKa = 11.84KKK47 pKa = 7.45KKK49 pKa = 9.95RR50 pKa = 11.84VAARR54 pKa = 11.84KKK56 pKa = 7.35KKK58 pKa = 9.9RR59 pKa = 11.84VSARR63 pKa = 11.84KKK65 pKa = 8.52KKK67 pKa = 10.04RR68 pKa = 11.84VAARR72 pKa = 11.84KKK74 pKa = 7.45KKK76 pKa = 9.95RR77 pKa = 11.84VAARR81 pKa = 11.84KKK83 pKa = 7.35KKK85 pKa = 9.9RR86 pKa = 11.84VSARR90 pKa = 11.84KKK92 pKa = 8.52KKK94 pKa = 10.04RR95 pKa = 11.84VAARR99 pKa = 11.84KKK101 pKa = 7.45KKK103 pKa = 9.95RR104 pKa = 11.84VAARR108 pKa = 11.84KKK110 pKa = 7.35KKK112 pKa = 9.9RR113 pKa = 11.84VSARR117 pKa = 11.84KKK119 pKa = 8.52KKK121 pKa = 10.04RR122 pKa = 11.84VAARR126 pKa = 11.84KKK128 pKa = 7.45KKK130 pKa = 9.95RR131 pKa = 11.84VAARR135 pKa = 11.84KKK137 pKa = 7.35KKK139 pKa = 9.9RR140 pKa = 11.84VSARR144 pKa = 11.84KKK146 pKa = 8.52KKK148 pKa = 10.04RR149 pKa = 11.84VAARR153 pKa = 11.84KKK155 pKa = 7.45KKK157 pKa = 9.95RR158 pKa = 11.84VAARR162 pKa = 11.84KKK164 pKa = 7.35KKK166 pKa = 9.9RR167 pKa = 11.84VSARR171 pKa = 11.84KKK173 pKa = 8.66KKK175 pKa = 9.76QYYY178 pKa = 10.11FKKK181 pKa = 10.61NPSVGPQL
Molecular weight: 21.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
24682227 |
22 |
7046 |
677.2 |
75.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.69 ± 0.012 | 2.309 ± 0.01 |
4.898 ± 0.008 | 7.111 ± 0.014 |
3.876 ± 0.009 | 6.138 ± 0.014 |
2.588 ± 0.006 | 4.852 ± 0.009 |
6.188 ± 0.015 | 9.867 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.267 ± 0.004 | 3.981 ± 0.009 |
5.516 ± 0.015 | 4.676 ± 0.011 |
5.472 ± 0.01 | 8.122 ± 0.015 |
5.296 ± 0.008 | 6.05 ± 0.009 |
1.266 ± 0.004 | 2.834 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
