
Pseudogulbenkiania sp. (strain NH8B)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Neisseriales; Chromobacteriaceae; Pseudogulbenkiania; unclassified Pseudogulbenkiania
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
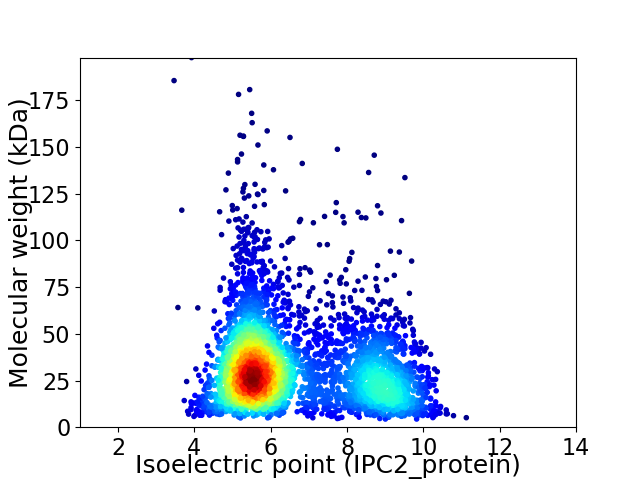
Virtual 2D-PAGE plot for 3958 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G2IWY2|G2IWY2_PSEUL 2-hydroxy-3-oxopropionate reductase OS=Pseudogulbenkiania sp. (strain NH8B) OX=748280 GN=NH8B_0496 PE=4 SV=1
MM1 pKa = 7.87ASTTTSGIITSFSNTPQAGDD21 pKa = 3.93DD22 pKa = 4.28LFTSSSTGLTEE33 pKa = 4.53DD34 pKa = 3.44SSSIVYY40 pKa = 10.08LNVMANDD47 pKa = 3.87LGGAAKK53 pKa = 9.36TLYY56 pKa = 10.85SVDD59 pKa = 4.57DD60 pKa = 4.11GTNSAGITSSTDD72 pKa = 3.37LLTKK76 pKa = 9.87DD77 pKa = 3.2TAYY80 pKa = 9.77STDD83 pKa = 3.24VSAHH87 pKa = 5.84GARR90 pKa = 11.84IWITTDD96 pKa = 2.48GRR98 pKa = 11.84VGYY101 pKa = 9.65DD102 pKa = 3.17ASTLSADD109 pKa = 5.21FKK111 pKa = 10.98ATLNALGAGEE121 pKa = 4.33YY122 pKa = 8.52QTDD125 pKa = 3.48TFTYY129 pKa = 10.19AIRR132 pKa = 11.84LGNGTLSWATATVQFAGVNDD152 pKa = 3.83AATFSGDD159 pKa = 3.36DD160 pKa = 3.56TKK162 pKa = 11.76GLTEE166 pKa = 3.9TDD168 pKa = 3.16AAQTTGGTLVAHH180 pKa = 6.83DD181 pKa = 4.2VDD183 pKa = 4.43SAEE186 pKa = 4.22TFQAQSNVAGSSGYY200 pKa = 10.14GHH202 pKa = 6.25FTIGSDD208 pKa = 3.86GTWSYY213 pKa = 11.28TMDD216 pKa = 3.53SAHH219 pKa = 6.83NEE221 pKa = 3.96FKK223 pKa = 11.14DD224 pKa = 3.51GVNYY228 pKa = 10.75SDD230 pKa = 3.49TLVVKK235 pKa = 10.54SADD238 pKa = 3.8GTEE241 pKa = 3.95HH242 pKa = 6.63TLTVNIAGTNDD253 pKa = 3.04AATFSGDD260 pKa = 3.38DD261 pKa = 3.64SKK263 pKa = 12.03GLTEE267 pKa = 3.9TDD269 pKa = 3.31AAQSTGGTLVAHH281 pKa = 6.83DD282 pKa = 4.2VDD284 pKa = 4.43SAEE287 pKa = 4.22TFQAQTNVAGSNGYY301 pKa = 8.51GHH303 pKa = 6.27FTIGSDD309 pKa = 3.86GTWSYY314 pKa = 11.28TMDD317 pKa = 3.53SAHH320 pKa = 6.9NEE322 pKa = 4.08FKK324 pKa = 10.97DD325 pKa = 3.42GEE327 pKa = 4.48NYY329 pKa = 10.63SDD331 pKa = 3.55TLVVQSADD339 pKa = 3.58GTEE342 pKa = 3.96HH343 pKa = 6.66TLTVNITGTNDD354 pKa = 2.93AATFTGDD361 pKa = 3.4DD362 pKa = 3.88SKK364 pKa = 11.97GLTEE368 pKa = 4.07TDD370 pKa = 3.03AAQTTGGTLVAHH382 pKa = 6.83DD383 pKa = 4.2VDD385 pKa = 4.43SAEE388 pKa = 4.22TFQAQTNVAGSNGYY402 pKa = 8.51GHH404 pKa = 6.27FTIGSDD410 pKa = 3.86GTWSYY415 pKa = 11.28TMDD418 pKa = 3.52SAHH421 pKa = 6.67NEE423 pKa = 3.84FAAGTTYY430 pKa = 11.33SDD432 pKa = 3.52TLVVQSADD440 pKa = 3.58GTEE443 pKa = 3.96HH444 pKa = 6.61TLTVNILGTNDD455 pKa = 3.14AATFTGDD462 pKa = 3.4DD463 pKa = 3.88SKK465 pKa = 11.97GLTEE469 pKa = 4.07TDD471 pKa = 3.03AAQTTGGTLVAHH483 pKa = 6.83DD484 pKa = 4.2VDD486 pKa = 4.43SAEE489 pKa = 4.22TFQAQSNVAGSNGYY503 pKa = 8.51GHH505 pKa = 6.27FTIGSDD511 pKa = 3.86GTWSYY516 pKa = 11.28TMDD519 pKa = 3.53SAHH522 pKa = 6.83NEE524 pKa = 3.96FKK526 pKa = 11.14DD527 pKa = 3.51GVNYY531 pKa = 10.75SDD533 pKa = 3.49TLVVKK538 pKa = 10.54SADD541 pKa = 3.8GTEE544 pKa = 3.95HH545 pKa = 6.68TLTVNITGTNDD556 pKa = 2.92AATFSGDD563 pKa = 3.38DD564 pKa = 3.64SKK566 pKa = 12.03GLTEE570 pKa = 3.9TDD572 pKa = 3.31AAQSTGGTLVAHH584 pKa = 6.83DD585 pKa = 4.2VDD587 pKa = 4.43SAEE590 pKa = 4.22TFQAQTNVAGSQGYY604 pKa = 7.93GHH606 pKa = 6.24FTIGSDD612 pKa = 4.0GAWSYY617 pKa = 11.29TMDD620 pKa = 3.6SAHH623 pKa = 6.83NEE625 pKa = 3.96FKK627 pKa = 11.11DD628 pKa = 3.57GVNYY632 pKa = 10.36TDD634 pKa = 3.48TLVVKK639 pKa = 10.48SADD642 pKa = 3.8GTEE645 pKa = 3.95HH646 pKa = 6.68TLTVNITGTNDD657 pKa = 2.92AATFSGDD664 pKa = 3.36DD665 pKa = 3.56TKK667 pKa = 11.76GLTEE671 pKa = 3.9TDD673 pKa = 3.16AAQTTGGTLVAHH685 pKa = 6.83DD686 pKa = 4.2VDD688 pKa = 4.43SAEE691 pKa = 4.22TFQAQTNVAGSQGYY705 pKa = 7.93GHH707 pKa = 6.24FTIGSDD713 pKa = 3.94GTWSYY718 pKa = 12.25SMDD721 pKa = 3.58SAHH724 pKa = 7.65DD725 pKa = 3.72EE726 pKa = 4.41FAGGTTNTDD735 pKa = 3.37TLVVQSADD743 pKa = 3.58GTEE746 pKa = 3.96HH747 pKa = 6.66TLTVNITGTNDD758 pKa = 3.03AATITASASEE768 pKa = 4.33DD769 pKa = 3.54TSVTEE774 pKa = 4.04VGSGVAGDD782 pKa = 4.15SSAGGQLTVHH792 pKa = 6.78DD793 pKa = 4.34VDD795 pKa = 3.46NGEE798 pKa = 4.39AHH800 pKa = 6.26FQTPVSLTGTYY811 pKa = 11.12GDD813 pKa = 4.1FSFNASTGVWSYY825 pKa = 12.0ALDD828 pKa = 3.78DD829 pKa = 4.05TRR831 pKa = 11.84PATDD835 pKa = 3.44ALATGSVVHH844 pKa = 6.69DD845 pKa = 3.72TLTVKK850 pKa = 10.72SFDD853 pKa = 3.39GSASYY858 pKa = 9.93TIDD861 pKa = 3.34VAVNGSNDD869 pKa = 3.9SVNHH873 pKa = 6.34APTDD877 pKa = 3.51IVLSMAEE884 pKa = 4.09PGGNALPGAGAVLGTLSTIDD904 pKa = 3.62VDD906 pKa = 4.32SGDD909 pKa = 3.55TFTYY913 pKa = 10.9SLISSSTVSGSAASFSISGNSLSTGTGLASNSVYY947 pKa = 10.62QLDD950 pKa = 3.69IQTTDD955 pKa = 3.14SASATYY961 pKa = 10.43HH962 pKa = 5.36EE963 pKa = 5.1VFNIITGTNAQGQVTGDD980 pKa = 4.03DD981 pKa = 3.78GSLPSGGGANSILIGDD997 pKa = 4.03DD998 pKa = 3.26VLYY1001 pKa = 10.99GSGGNDD1007 pKa = 3.05VMFGGSGNDD1016 pKa = 3.31TLFGQNGNDD1025 pKa = 3.43VLYY1028 pKa = 10.92GGAGNDD1034 pKa = 3.65TLSGGNQADD1043 pKa = 3.37RR1044 pKa = 11.84FVFLASDD1051 pKa = 3.32NGGTDD1056 pKa = 3.64TITNFTATGANADD1069 pKa = 4.01TLDD1072 pKa = 4.77ISDD1075 pKa = 5.32LLINYY1080 pKa = 8.58NPATPSAFVHH1090 pKa = 5.75FTEE1093 pKa = 5.14SGGNTIVSVDD1103 pKa = 3.47RR1104 pKa = 11.84DD1105 pKa = 3.83GSGGSYY1111 pKa = 10.43GFQDD1115 pKa = 3.47VAILNGVTGLNMNDD1129 pKa = 3.23MLTSGKK1135 pKa = 10.29LDD1137 pKa = 3.45VVPP1140 pKa = 5.06
MM1 pKa = 7.87ASTTTSGIITSFSNTPQAGDD21 pKa = 3.93DD22 pKa = 4.28LFTSSSTGLTEE33 pKa = 4.53DD34 pKa = 3.44SSSIVYY40 pKa = 10.08LNVMANDD47 pKa = 3.87LGGAAKK53 pKa = 9.36TLYY56 pKa = 10.85SVDD59 pKa = 4.57DD60 pKa = 4.11GTNSAGITSSTDD72 pKa = 3.37LLTKK76 pKa = 9.87DD77 pKa = 3.2TAYY80 pKa = 9.77STDD83 pKa = 3.24VSAHH87 pKa = 5.84GARR90 pKa = 11.84IWITTDD96 pKa = 2.48GRR98 pKa = 11.84VGYY101 pKa = 9.65DD102 pKa = 3.17ASTLSADD109 pKa = 5.21FKK111 pKa = 10.98ATLNALGAGEE121 pKa = 4.33YY122 pKa = 8.52QTDD125 pKa = 3.48TFTYY129 pKa = 10.19AIRR132 pKa = 11.84LGNGTLSWATATVQFAGVNDD152 pKa = 3.83AATFSGDD159 pKa = 3.36DD160 pKa = 3.56TKK162 pKa = 11.76GLTEE166 pKa = 3.9TDD168 pKa = 3.16AAQTTGGTLVAHH180 pKa = 6.83DD181 pKa = 4.2VDD183 pKa = 4.43SAEE186 pKa = 4.22TFQAQSNVAGSSGYY200 pKa = 10.14GHH202 pKa = 6.25FTIGSDD208 pKa = 3.86GTWSYY213 pKa = 11.28TMDD216 pKa = 3.53SAHH219 pKa = 6.83NEE221 pKa = 3.96FKK223 pKa = 11.14DD224 pKa = 3.51GVNYY228 pKa = 10.75SDD230 pKa = 3.49TLVVKK235 pKa = 10.54SADD238 pKa = 3.8GTEE241 pKa = 3.95HH242 pKa = 6.63TLTVNIAGTNDD253 pKa = 3.04AATFSGDD260 pKa = 3.38DD261 pKa = 3.64SKK263 pKa = 12.03GLTEE267 pKa = 3.9TDD269 pKa = 3.31AAQSTGGTLVAHH281 pKa = 6.83DD282 pKa = 4.2VDD284 pKa = 4.43SAEE287 pKa = 4.22TFQAQTNVAGSNGYY301 pKa = 8.51GHH303 pKa = 6.27FTIGSDD309 pKa = 3.86GTWSYY314 pKa = 11.28TMDD317 pKa = 3.53SAHH320 pKa = 6.9NEE322 pKa = 4.08FKK324 pKa = 10.97DD325 pKa = 3.42GEE327 pKa = 4.48NYY329 pKa = 10.63SDD331 pKa = 3.55TLVVQSADD339 pKa = 3.58GTEE342 pKa = 3.96HH343 pKa = 6.66TLTVNITGTNDD354 pKa = 2.93AATFTGDD361 pKa = 3.4DD362 pKa = 3.88SKK364 pKa = 11.97GLTEE368 pKa = 4.07TDD370 pKa = 3.03AAQTTGGTLVAHH382 pKa = 6.83DD383 pKa = 4.2VDD385 pKa = 4.43SAEE388 pKa = 4.22TFQAQTNVAGSNGYY402 pKa = 8.51GHH404 pKa = 6.27FTIGSDD410 pKa = 3.86GTWSYY415 pKa = 11.28TMDD418 pKa = 3.52SAHH421 pKa = 6.67NEE423 pKa = 3.84FAAGTTYY430 pKa = 11.33SDD432 pKa = 3.52TLVVQSADD440 pKa = 3.58GTEE443 pKa = 3.96HH444 pKa = 6.61TLTVNILGTNDD455 pKa = 3.14AATFTGDD462 pKa = 3.4DD463 pKa = 3.88SKK465 pKa = 11.97GLTEE469 pKa = 4.07TDD471 pKa = 3.03AAQTTGGTLVAHH483 pKa = 6.83DD484 pKa = 4.2VDD486 pKa = 4.43SAEE489 pKa = 4.22TFQAQSNVAGSNGYY503 pKa = 8.51GHH505 pKa = 6.27FTIGSDD511 pKa = 3.86GTWSYY516 pKa = 11.28TMDD519 pKa = 3.53SAHH522 pKa = 6.83NEE524 pKa = 3.96FKK526 pKa = 11.14DD527 pKa = 3.51GVNYY531 pKa = 10.75SDD533 pKa = 3.49TLVVKK538 pKa = 10.54SADD541 pKa = 3.8GTEE544 pKa = 3.95HH545 pKa = 6.68TLTVNITGTNDD556 pKa = 2.92AATFSGDD563 pKa = 3.38DD564 pKa = 3.64SKK566 pKa = 12.03GLTEE570 pKa = 3.9TDD572 pKa = 3.31AAQSTGGTLVAHH584 pKa = 6.83DD585 pKa = 4.2VDD587 pKa = 4.43SAEE590 pKa = 4.22TFQAQTNVAGSQGYY604 pKa = 7.93GHH606 pKa = 6.24FTIGSDD612 pKa = 4.0GAWSYY617 pKa = 11.29TMDD620 pKa = 3.6SAHH623 pKa = 6.83NEE625 pKa = 3.96FKK627 pKa = 11.11DD628 pKa = 3.57GVNYY632 pKa = 10.36TDD634 pKa = 3.48TLVVKK639 pKa = 10.48SADD642 pKa = 3.8GTEE645 pKa = 3.95HH646 pKa = 6.68TLTVNITGTNDD657 pKa = 2.92AATFSGDD664 pKa = 3.36DD665 pKa = 3.56TKK667 pKa = 11.76GLTEE671 pKa = 3.9TDD673 pKa = 3.16AAQTTGGTLVAHH685 pKa = 6.83DD686 pKa = 4.2VDD688 pKa = 4.43SAEE691 pKa = 4.22TFQAQTNVAGSQGYY705 pKa = 7.93GHH707 pKa = 6.24FTIGSDD713 pKa = 3.94GTWSYY718 pKa = 12.25SMDD721 pKa = 3.58SAHH724 pKa = 7.65DD725 pKa = 3.72EE726 pKa = 4.41FAGGTTNTDD735 pKa = 3.37TLVVQSADD743 pKa = 3.58GTEE746 pKa = 3.96HH747 pKa = 6.66TLTVNITGTNDD758 pKa = 3.03AATITASASEE768 pKa = 4.33DD769 pKa = 3.54TSVTEE774 pKa = 4.04VGSGVAGDD782 pKa = 4.15SSAGGQLTVHH792 pKa = 6.78DD793 pKa = 4.34VDD795 pKa = 3.46NGEE798 pKa = 4.39AHH800 pKa = 6.26FQTPVSLTGTYY811 pKa = 11.12GDD813 pKa = 4.1FSFNASTGVWSYY825 pKa = 12.0ALDD828 pKa = 3.78DD829 pKa = 4.05TRR831 pKa = 11.84PATDD835 pKa = 3.44ALATGSVVHH844 pKa = 6.69DD845 pKa = 3.72TLTVKK850 pKa = 10.72SFDD853 pKa = 3.39GSASYY858 pKa = 9.93TIDD861 pKa = 3.34VAVNGSNDD869 pKa = 3.9SVNHH873 pKa = 6.34APTDD877 pKa = 3.51IVLSMAEE884 pKa = 4.09PGGNALPGAGAVLGTLSTIDD904 pKa = 3.62VDD906 pKa = 4.32SGDD909 pKa = 3.55TFTYY913 pKa = 10.9SLISSSTVSGSAASFSISGNSLSTGTGLASNSVYY947 pKa = 10.62QLDD950 pKa = 3.69IQTTDD955 pKa = 3.14SASATYY961 pKa = 10.43HH962 pKa = 5.36EE963 pKa = 5.1VFNIITGTNAQGQVTGDD980 pKa = 4.03DD981 pKa = 3.78GSLPSGGGANSILIGDD997 pKa = 4.03DD998 pKa = 3.26VLYY1001 pKa = 10.99GSGGNDD1007 pKa = 3.05VMFGGSGNDD1016 pKa = 3.31TLFGQNGNDD1025 pKa = 3.43VLYY1028 pKa = 10.92GGAGNDD1034 pKa = 3.65TLSGGNQADD1043 pKa = 3.37RR1044 pKa = 11.84FVFLASDD1051 pKa = 3.32NGGTDD1056 pKa = 3.64TITNFTATGANADD1069 pKa = 4.01TLDD1072 pKa = 4.77ISDD1075 pKa = 5.32LLINYY1080 pKa = 8.58NPATPSAFVHH1090 pKa = 5.75FTEE1093 pKa = 5.14SGGNTIVSVDD1103 pKa = 3.47RR1104 pKa = 11.84DD1105 pKa = 3.83GSGGSYY1111 pKa = 10.43GFQDD1115 pKa = 3.47VAILNGVTGLNMNDD1129 pKa = 3.23MLTSGKK1135 pKa = 10.29LDD1137 pKa = 3.45VVPP1140 pKa = 5.06
Molecular weight: 116.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G2J349|G2J349_PSEUL Uncharacterized protein OS=Pseudogulbenkiania sp. (strain NH8B) OX=748280 GN=NH8B_1677 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.31QPSTTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.24RR14 pKa = 11.84THH16 pKa = 5.92GFLVRR21 pKa = 11.84SKK23 pKa = 9.38TRR25 pKa = 11.84GGRR28 pKa = 11.84AVLAARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.45GRR39 pKa = 11.84KK40 pKa = 8.54RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.4RR3 pKa = 11.84TYY5 pKa = 10.31QPSTTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.24RR14 pKa = 11.84THH16 pKa = 5.92GFLVRR21 pKa = 11.84SKK23 pKa = 9.38TRR25 pKa = 11.84GGRR28 pKa = 11.84AVLAARR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.45GRR39 pKa = 11.84KK40 pKa = 8.54RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1248374 |
40 |
1991 |
315.4 |
34.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.907 ± 0.052 | 0.973 ± 0.013 |
5.251 ± 0.026 | 5.717 ± 0.04 |
3.572 ± 0.024 | 8.077 ± 0.036 |
2.367 ± 0.02 | 4.58 ± 0.031 |
3.492 ± 0.036 | 11.533 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.372 ± 0.016 | 2.738 ± 0.025 |
5.012 ± 0.027 | 4.089 ± 0.031 |
6.659 ± 0.043 | 5.442 ± 0.031 |
4.909 ± 0.034 | 7.398 ± 0.033 |
1.423 ± 0.02 | 2.489 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
