
Nocardioides sp. Root151
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
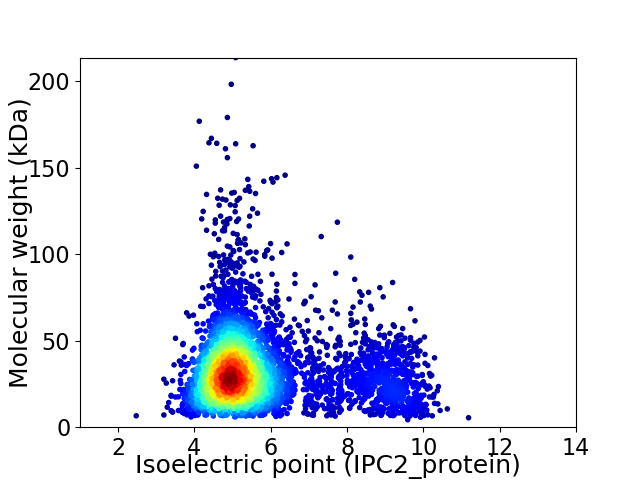
Virtual 2D-PAGE plot for 4609 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q7YYE7|A0A0Q7YYE7_9ACTN Uncharacterized protein OS=Nocardioides sp. Root151 OX=1736475 GN=ASD66_17380 PE=4 SV=1
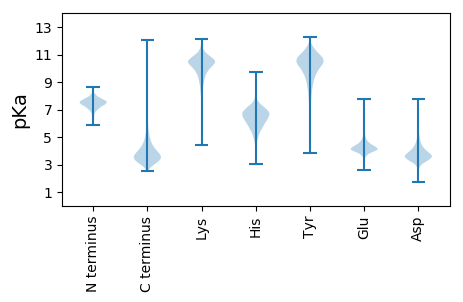
MM1 pKa = 7.62PGSATALSGGATVDD15 pKa = 3.65PQPAALADD23 pKa = 4.05APDD26 pKa = 3.91IPLSAVTAHH35 pKa = 6.83LEE37 pKa = 4.11EE38 pKa = 4.49FQSIAEE44 pKa = 4.12ANGGNRR50 pKa = 11.84YY51 pKa = 7.69TGQPGYY57 pKa = 9.87RR58 pKa = 11.84ASLDD62 pKa = 3.62YY63 pKa = 11.31VQAQLDD69 pKa = 3.58AAGFVTEE76 pKa = 4.24VQSFSTRR83 pKa = 11.84AGTSYY88 pKa = 11.52NLIADD93 pKa = 4.15WPGGDD98 pKa = 3.65PTNTVMMGGHH108 pKa = 7.3LDD110 pKa = 3.83SVDD113 pKa = 3.77EE114 pKa = 4.69GPGINDD120 pKa = 3.58NGTGSAAILASALAFAQSGAEE141 pKa = 4.05PTNHH145 pKa = 5.17VRR147 pKa = 11.84FGFWGAEE154 pKa = 3.85EE155 pKa = 4.8LGLLGSTHH163 pKa = 6.15YY164 pKa = 10.84VDD166 pKa = 4.09SLSTVEE172 pKa = 4.29VDD174 pKa = 5.21DD175 pKa = 4.1LAAYY179 pKa = 10.0FNFDD183 pKa = 3.26MVGSPNPGYY192 pKa = 10.09FVYY195 pKa = 10.76DD196 pKa = 3.54DD197 pKa = 4.6DD198 pKa = 6.65ANGTEE203 pKa = 4.79LRR205 pKa = 11.84DD206 pKa = 3.84SLTASYY212 pKa = 10.51DD213 pKa = 3.32AAGIDD218 pKa = 3.6SDD220 pKa = 4.85YY221 pKa = 11.09IDD223 pKa = 3.28VDD225 pKa = 3.49GRR227 pKa = 11.84SDD229 pKa = 3.03HH230 pKa = 6.61AAFMQRR236 pKa = 11.84GIPTAGTFSGAEE248 pKa = 4.06GTKK251 pKa = 10.12TSTQAAKK258 pKa = 9.91WGGQAGQAYY267 pKa = 9.24DD268 pKa = 3.6PCYY271 pKa = 10.54HH272 pKa = 6.43SACDD276 pKa = 5.79DD277 pKa = 3.48IDD279 pKa = 4.31NLDD282 pKa = 3.67TAALDD287 pKa = 4.05LNADD291 pKa = 5.01LIADD295 pKa = 4.41TLWAWADD302 pKa = 3.16HH303 pKa = 6.99DD304 pKa = 4.44FGGGTTEE311 pKa = 4.92PPTGDD316 pKa = 3.91DD317 pKa = 4.45AITNGGFEE325 pKa = 5.28DD326 pKa = 5.28GTTGWQATNGVVTTDD341 pKa = 4.23LDD343 pKa = 3.75QSAHH347 pKa = 5.91GGSGKK352 pKa = 10.44AWLNGYY358 pKa = 8.88GRR360 pKa = 11.84AHH362 pKa = 6.8TDD364 pKa = 2.84SVAQTVTVPDD374 pKa = 3.83AGRR377 pKa = 11.84LTFWLRR383 pKa = 11.84VDD385 pKa = 3.51TDD387 pKa = 3.37EE388 pKa = 4.83DD389 pKa = 4.32TTGQAYY395 pKa = 7.84DD396 pKa = 3.67TLRR399 pKa = 11.84VKK401 pKa = 10.79AGSTTLGTWSNLDD414 pKa = 3.44AGDD417 pKa = 3.85YY418 pKa = 10.74RR419 pKa = 11.84SIALDD424 pKa = 3.69LSAYY428 pKa = 9.9AGQSVALTFVGTEE441 pKa = 3.98DD442 pKa = 4.52AYY444 pKa = 11.18LASDD448 pKa = 3.8FVIDD452 pKa = 5.04DD453 pKa = 3.76VSLTSAGG460 pKa = 3.5
MM1 pKa = 7.62PGSATALSGGATVDD15 pKa = 3.65PQPAALADD23 pKa = 4.05APDD26 pKa = 3.91IPLSAVTAHH35 pKa = 6.83LEE37 pKa = 4.11EE38 pKa = 4.49FQSIAEE44 pKa = 4.12ANGGNRR50 pKa = 11.84YY51 pKa = 7.69TGQPGYY57 pKa = 9.87RR58 pKa = 11.84ASLDD62 pKa = 3.62YY63 pKa = 11.31VQAQLDD69 pKa = 3.58AAGFVTEE76 pKa = 4.24VQSFSTRR83 pKa = 11.84AGTSYY88 pKa = 11.52NLIADD93 pKa = 4.15WPGGDD98 pKa = 3.65PTNTVMMGGHH108 pKa = 7.3LDD110 pKa = 3.83SVDD113 pKa = 3.77EE114 pKa = 4.69GPGINDD120 pKa = 3.58NGTGSAAILASALAFAQSGAEE141 pKa = 4.05PTNHH145 pKa = 5.17VRR147 pKa = 11.84FGFWGAEE154 pKa = 3.85EE155 pKa = 4.8LGLLGSTHH163 pKa = 6.15YY164 pKa = 10.84VDD166 pKa = 4.09SLSTVEE172 pKa = 4.29VDD174 pKa = 5.21DD175 pKa = 4.1LAAYY179 pKa = 10.0FNFDD183 pKa = 3.26MVGSPNPGYY192 pKa = 10.09FVYY195 pKa = 10.76DD196 pKa = 3.54DD197 pKa = 4.6DD198 pKa = 6.65ANGTEE203 pKa = 4.79LRR205 pKa = 11.84DD206 pKa = 3.84SLTASYY212 pKa = 10.51DD213 pKa = 3.32AAGIDD218 pKa = 3.6SDD220 pKa = 4.85YY221 pKa = 11.09IDD223 pKa = 3.28VDD225 pKa = 3.49GRR227 pKa = 11.84SDD229 pKa = 3.03HH230 pKa = 6.61AAFMQRR236 pKa = 11.84GIPTAGTFSGAEE248 pKa = 4.06GTKK251 pKa = 10.12TSTQAAKK258 pKa = 9.91WGGQAGQAYY267 pKa = 9.24DD268 pKa = 3.6PCYY271 pKa = 10.54HH272 pKa = 6.43SACDD276 pKa = 5.79DD277 pKa = 3.48IDD279 pKa = 4.31NLDD282 pKa = 3.67TAALDD287 pKa = 4.05LNADD291 pKa = 5.01LIADD295 pKa = 4.41TLWAWADD302 pKa = 3.16HH303 pKa = 6.99DD304 pKa = 4.44FGGGTTEE311 pKa = 4.92PPTGDD316 pKa = 3.91DD317 pKa = 4.45AITNGGFEE325 pKa = 5.28DD326 pKa = 5.28GTTGWQATNGVVTTDD341 pKa = 4.23LDD343 pKa = 3.75QSAHH347 pKa = 5.91GGSGKK352 pKa = 10.44AWLNGYY358 pKa = 8.88GRR360 pKa = 11.84AHH362 pKa = 6.8TDD364 pKa = 2.84SVAQTVTVPDD374 pKa = 3.83AGRR377 pKa = 11.84LTFWLRR383 pKa = 11.84VDD385 pKa = 3.51TDD387 pKa = 3.37EE388 pKa = 4.83DD389 pKa = 4.32TTGQAYY395 pKa = 7.84DD396 pKa = 3.67TLRR399 pKa = 11.84VKK401 pKa = 10.79AGSTTLGTWSNLDD414 pKa = 3.44AGDD417 pKa = 3.85YY418 pKa = 10.74RR419 pKa = 11.84SIALDD424 pKa = 3.69LSAYY428 pKa = 9.9AGQSVALTFVGTEE441 pKa = 3.98DD442 pKa = 4.52AYY444 pKa = 11.18LASDD448 pKa = 3.8FVIDD452 pKa = 5.04DD453 pKa = 3.76VSLTSAGG460 pKa = 3.5
Molecular weight: 47.73 kDa
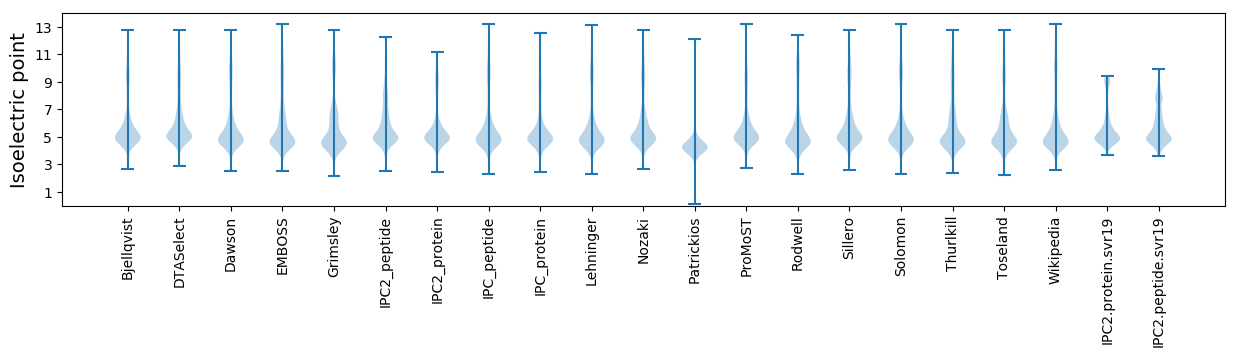
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q7Z7H2|A0A0Q7Z7H2_9ACTN Probable nicotinate-nucleotide adenylyltransferase OS=Nocardioides sp. Root151 OX=1736475 GN=nadD PE=3 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 4.97KK15 pKa = 10.04VHH17 pKa = 6.59GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSTRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.87GRR40 pKa = 11.84SKK42 pKa = 10.84LAVV45 pKa = 3.25
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 4.97KK15 pKa = 10.04VHH17 pKa = 6.59GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSTRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.87GRR40 pKa = 11.84SKK42 pKa = 10.84LAVV45 pKa = 3.25
Molecular weight: 5.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1475871 |
37 |
1974 |
320.2 |
34.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.45 ± 0.045 | 0.738 ± 0.009 |
6.726 ± 0.03 | 5.867 ± 0.034 |
2.974 ± 0.019 | 9.079 ± 0.034 |
2.281 ± 0.019 | 3.784 ± 0.024 |
2.333 ± 0.025 | 10.128 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.004 ± 0.014 | 2.03 ± 0.018 |
5.345 ± 0.023 | 2.767 ± 0.017 |
7.19 ± 0.04 | 5.482 ± 0.02 |
6.181 ± 0.032 | 9.18 ± 0.032 |
1.512 ± 0.017 | 1.946 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
