
Kemerovo virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus; Great Island virus
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
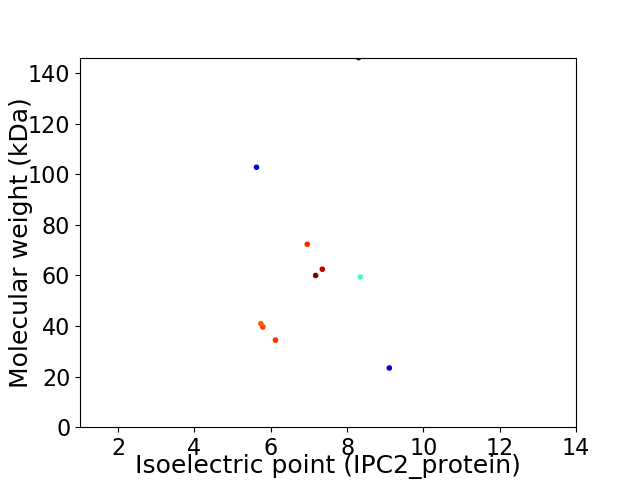
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M4MJR4|M4MJR4_9REOV Non-structural protein NS2 OS=Kemerovo virus OX=40064 GN=NS2 PE=4 SV=1
MM1 pKa = 7.61SSDD4 pKa = 3.35KK5 pKa = 11.22QNVPMIKK12 pKa = 10.12KK13 pKa = 8.89PFTRR17 pKa = 11.84VIVLKK22 pKa = 10.83SDD24 pKa = 3.09GRR26 pKa = 11.84DD27 pKa = 3.28DD28 pKa = 5.59FVAKK32 pKa = 9.62MCAVLGCEE40 pKa = 4.12YY41 pKa = 10.94VVVRR45 pKa = 11.84HH46 pKa = 6.02GLGTQIAGSDD56 pKa = 3.92TPAHH60 pKa = 6.21GCLLLTIPGPGSFRR74 pKa = 11.84MIDD77 pKa = 3.36RR78 pKa = 11.84DD79 pKa = 3.26QSTFLVVSGDD89 pKa = 3.62GLEE92 pKa = 4.21VQQDD96 pKa = 3.09RR97 pKa = 11.84WTGMRR102 pKa = 11.84FEE104 pKa = 4.77AVDD107 pKa = 3.67MYY109 pKa = 11.12PRR111 pKa = 11.84ACRR114 pKa = 11.84IRR116 pKa = 11.84YY117 pKa = 8.79GEE119 pKa = 4.44KK120 pKa = 10.66EE121 pKa = 3.91NDD123 pKa = 3.08SEE125 pKa = 4.39IRR127 pKa = 11.84FGRR130 pKa = 11.84ASGTVPPYY138 pKa = 10.31TPDD141 pKa = 3.3GVSEE145 pKa = 4.11EE146 pKa = 4.19QEE148 pKa = 4.18EE149 pKa = 4.53VALPGISFHH158 pKa = 6.26TPEE161 pKa = 4.57EE162 pKa = 4.42DD163 pKa = 3.3YY164 pKa = 11.35KK165 pKa = 11.01EE166 pKa = 3.86YY167 pKa = 10.64RR168 pKa = 11.84EE169 pKa = 4.02RR170 pKa = 11.84LRR172 pKa = 11.84EE173 pKa = 3.94EE174 pKa = 3.92RR175 pKa = 11.84DD176 pKa = 3.35LRR178 pKa = 11.84SANILEE184 pKa = 4.44ALRR187 pKa = 11.84TTHH190 pKa = 6.65QSRR193 pKa = 11.84TLGRR197 pKa = 11.84IHH199 pKa = 7.93GIRR202 pKa = 11.84NSEE205 pKa = 4.21LTTLRR210 pKa = 11.84PPAPPTLRR218 pKa = 11.84RR219 pKa = 11.84GVSSSNEE226 pKa = 3.48RR227 pKa = 11.84SADD230 pKa = 3.9RR231 pKa = 11.84TPTPPKK237 pKa = 9.37LTAPTYY243 pKa = 7.38PTAAAPRR250 pKa = 11.84AAAAPPPIPSVKK262 pKa = 9.75RR263 pKa = 11.84VSKK266 pKa = 8.77TAPQEE271 pKa = 4.54AEE273 pKa = 4.11SQCPKK278 pKa = 9.5FTHH281 pKa = 6.85EE282 pKa = 3.99YY283 pKa = 10.19DD284 pKa = 3.51QLAEE288 pKa = 4.06RR289 pKa = 11.84AFEE292 pKa = 4.77SLLEE296 pKa = 4.62DD297 pKa = 4.2DD298 pKa = 4.14PNARR302 pKa = 11.84FSYY305 pKa = 10.14GGAPEE310 pKa = 4.2TVGLFSQCLGTYY322 pKa = 9.44EE323 pKa = 4.66VDD325 pKa = 3.52PLMLPAYY332 pKa = 9.87EE333 pKa = 4.48IDD335 pKa = 3.55QTKK338 pKa = 10.21NRR340 pKa = 11.84YY341 pKa = 9.37KK342 pKa = 10.14YY343 pKa = 11.06VGMKK347 pKa = 9.74TSSQLNALVCDD358 pKa = 3.73DD359 pKa = 3.65RR360 pKa = 11.84VYY362 pKa = 10.72FVPSANN368 pKa = 3.3
MM1 pKa = 7.61SSDD4 pKa = 3.35KK5 pKa = 11.22QNVPMIKK12 pKa = 10.12KK13 pKa = 8.89PFTRR17 pKa = 11.84VIVLKK22 pKa = 10.83SDD24 pKa = 3.09GRR26 pKa = 11.84DD27 pKa = 3.28DD28 pKa = 5.59FVAKK32 pKa = 9.62MCAVLGCEE40 pKa = 4.12YY41 pKa = 10.94VVVRR45 pKa = 11.84HH46 pKa = 6.02GLGTQIAGSDD56 pKa = 3.92TPAHH60 pKa = 6.21GCLLLTIPGPGSFRR74 pKa = 11.84MIDD77 pKa = 3.36RR78 pKa = 11.84DD79 pKa = 3.26QSTFLVVSGDD89 pKa = 3.62GLEE92 pKa = 4.21VQQDD96 pKa = 3.09RR97 pKa = 11.84WTGMRR102 pKa = 11.84FEE104 pKa = 4.77AVDD107 pKa = 3.67MYY109 pKa = 11.12PRR111 pKa = 11.84ACRR114 pKa = 11.84IRR116 pKa = 11.84YY117 pKa = 8.79GEE119 pKa = 4.44KK120 pKa = 10.66EE121 pKa = 3.91NDD123 pKa = 3.08SEE125 pKa = 4.39IRR127 pKa = 11.84FGRR130 pKa = 11.84ASGTVPPYY138 pKa = 10.31TPDD141 pKa = 3.3GVSEE145 pKa = 4.11EE146 pKa = 4.19QEE148 pKa = 4.18EE149 pKa = 4.53VALPGISFHH158 pKa = 6.26TPEE161 pKa = 4.57EE162 pKa = 4.42DD163 pKa = 3.3YY164 pKa = 11.35KK165 pKa = 11.01EE166 pKa = 3.86YY167 pKa = 10.64RR168 pKa = 11.84EE169 pKa = 4.02RR170 pKa = 11.84LRR172 pKa = 11.84EE173 pKa = 3.94EE174 pKa = 3.92RR175 pKa = 11.84DD176 pKa = 3.35LRR178 pKa = 11.84SANILEE184 pKa = 4.44ALRR187 pKa = 11.84TTHH190 pKa = 6.65QSRR193 pKa = 11.84TLGRR197 pKa = 11.84IHH199 pKa = 7.93GIRR202 pKa = 11.84NSEE205 pKa = 4.21LTTLRR210 pKa = 11.84PPAPPTLRR218 pKa = 11.84RR219 pKa = 11.84GVSSSNEE226 pKa = 3.48RR227 pKa = 11.84SADD230 pKa = 3.9RR231 pKa = 11.84TPTPPKK237 pKa = 9.37LTAPTYY243 pKa = 7.38PTAAAPRR250 pKa = 11.84AAAAPPPIPSVKK262 pKa = 9.75RR263 pKa = 11.84VSKK266 pKa = 8.77TAPQEE271 pKa = 4.54AEE273 pKa = 4.11SQCPKK278 pKa = 9.5FTHH281 pKa = 6.85EE282 pKa = 3.99YY283 pKa = 10.19DD284 pKa = 3.51QLAEE288 pKa = 4.06RR289 pKa = 11.84AFEE292 pKa = 4.77SLLEE296 pKa = 4.62DD297 pKa = 4.2DD298 pKa = 4.14PNARR302 pKa = 11.84FSYY305 pKa = 10.14GGAPEE310 pKa = 4.2TVGLFSQCLGTYY322 pKa = 9.44EE323 pKa = 4.66VDD325 pKa = 3.52PLMLPAYY332 pKa = 9.87EE333 pKa = 4.48IDD335 pKa = 3.55QTKK338 pKa = 10.21NRR340 pKa = 11.84YY341 pKa = 9.37KK342 pKa = 10.14YY343 pKa = 11.06VGMKK347 pKa = 9.74TSSQLNALVCDD358 pKa = 3.73DD359 pKa = 3.65RR360 pKa = 11.84VYY362 pKa = 10.72FVPSANN368 pKa = 3.3
Molecular weight: 40.91 kDa
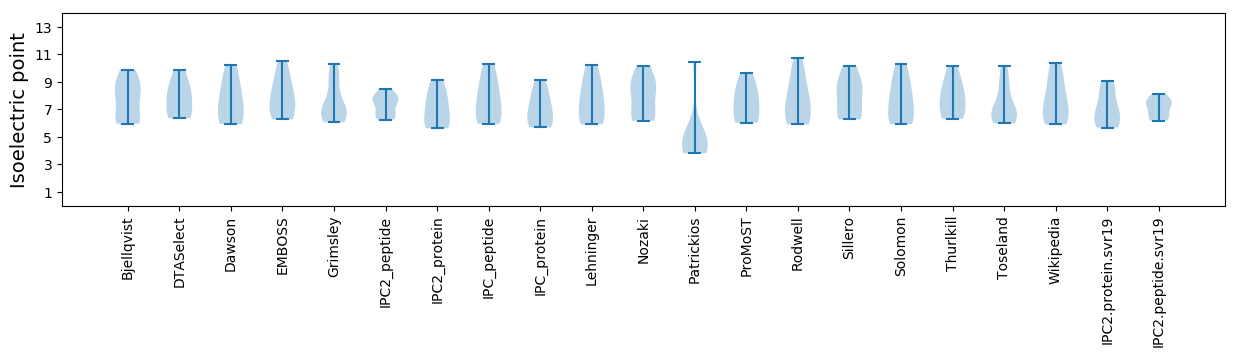
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M4MBL5|M4MBL5_9REOV RNA-directed RNA polymerase OS=Kemerovo virus OX=40064 GN=VP1 PE=3 SV=1
MM1 pKa = 7.47LAALEE6 pKa = 4.48MKK8 pKa = 10.27QSPTAPPAYY17 pKa = 9.92AATPMANVALGVLQNAITSTTGANEE42 pKa = 3.67TARR45 pKa = 11.84NEE47 pKa = 3.88KK48 pKa = 9.91AAYY51 pKa = 8.47GAAVEE56 pKa = 4.23ALKK59 pKa = 10.8DD60 pKa = 3.81DD61 pKa = 3.75EE62 pKa = 4.51TTRR65 pKa = 11.84MLKK68 pKa = 10.32AHH70 pKa = 5.8VNEE73 pKa = 4.24TSLRR77 pKa = 11.84EE78 pKa = 4.19AEE80 pKa = 4.02RR81 pKa = 11.84TLAKK85 pKa = 10.37LKK87 pKa = 10.52RR88 pKa = 11.84KK89 pKa = 9.6CLLLRR94 pKa = 11.84CAEE97 pKa = 4.17LMCLALTIGISVVLMITSAADD118 pKa = 3.19ALEE121 pKa = 4.38AQLKK125 pKa = 8.58RR126 pKa = 11.84INMSSHH132 pKa = 6.85VITGLLTAAAIGLQRR147 pKa = 11.84ARR149 pKa = 11.84QSHH152 pKa = 4.05QRR154 pKa = 11.84RR155 pKa = 11.84KK156 pKa = 10.34KK157 pKa = 9.26SLKK160 pKa = 9.23RR161 pKa = 11.84DD162 pKa = 3.55LVKK165 pKa = 10.82KK166 pKa = 10.0QMYY169 pKa = 8.86VSFVRR174 pKa = 11.84RR175 pKa = 11.84MGSQIPEE182 pKa = 3.96SSAAGPDD189 pKa = 3.23FHH191 pKa = 8.25ARR193 pKa = 11.84LLALAEE199 pKa = 3.73EE200 pKa = 4.62AGRR203 pKa = 11.84HH204 pKa = 5.48KK205 pKa = 10.04EE206 pKa = 4.16TSDD209 pKa = 2.83WRR211 pKa = 11.84HH212 pKa = 4.71WPP214 pKa = 3.35
MM1 pKa = 7.47LAALEE6 pKa = 4.48MKK8 pKa = 10.27QSPTAPPAYY17 pKa = 9.92AATPMANVALGVLQNAITSTTGANEE42 pKa = 3.67TARR45 pKa = 11.84NEE47 pKa = 3.88KK48 pKa = 9.91AAYY51 pKa = 8.47GAAVEE56 pKa = 4.23ALKK59 pKa = 10.8DD60 pKa = 3.81DD61 pKa = 3.75EE62 pKa = 4.51TTRR65 pKa = 11.84MLKK68 pKa = 10.32AHH70 pKa = 5.8VNEE73 pKa = 4.24TSLRR77 pKa = 11.84EE78 pKa = 4.19AEE80 pKa = 4.02RR81 pKa = 11.84TLAKK85 pKa = 10.37LKK87 pKa = 10.52RR88 pKa = 11.84KK89 pKa = 9.6CLLLRR94 pKa = 11.84CAEE97 pKa = 4.17LMCLALTIGISVVLMITSAADD118 pKa = 3.19ALEE121 pKa = 4.38AQLKK125 pKa = 8.58RR126 pKa = 11.84INMSSHH132 pKa = 6.85VITGLLTAAAIGLQRR147 pKa = 11.84ARR149 pKa = 11.84QSHH152 pKa = 4.05QRR154 pKa = 11.84RR155 pKa = 11.84KK156 pKa = 10.34KK157 pKa = 9.26SLKK160 pKa = 9.23RR161 pKa = 11.84DD162 pKa = 3.55LVKK165 pKa = 10.82KK166 pKa = 10.0QMYY169 pKa = 8.86VSFVRR174 pKa = 11.84RR175 pKa = 11.84MGSQIPEE182 pKa = 3.96SSAAGPDD189 pKa = 3.23FHH191 pKa = 8.25ARR193 pKa = 11.84LLALAEE199 pKa = 3.73EE200 pKa = 4.62AGRR203 pKa = 11.84HH204 pKa = 5.48KK205 pKa = 10.04EE206 pKa = 4.16TSDD209 pKa = 2.83WRR211 pKa = 11.84HH212 pKa = 4.71WPP214 pKa = 3.35
Molecular weight: 23.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5701 |
214 |
1285 |
570.1 |
64.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.963 ± 0.627 | 1.421 ± 0.312 |
5.911 ± 0.421 | 6.209 ± 0.363 |
3.491 ± 0.269 | 5.736 ± 0.328 |
3.298 ± 0.284 | 5.017 ± 0.379 |
2.842 ± 0.501 | 9.174 ± 0.392 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.561 ± 0.216 | 2.789 ± 0.145 |
5.297 ± 0.499 | 3.508 ± 0.431 |
9.577 ± 0.234 | 6.122 ± 0.27 |
6.157 ± 0.383 | 6.771 ± 0.214 |
1.052 ± 0.188 | 3.105 ± 0.269 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
