
Leishmania RNA virus 2 - 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Leishmaniavirus; Leishmania RNA virus 2
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins
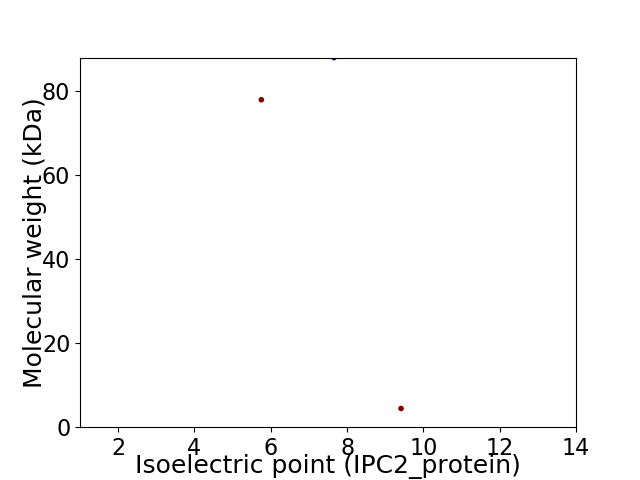
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q83073|Q83073_9VIRU RNA-directed RNA polymerase OS=Leishmania RNA virus 2 - 1 OX=39116 PE=3 SV=1
MM1 pKa = 7.58TSPIPQMNTSVFNLTRR17 pKa = 11.84GRR19 pKa = 11.84RR20 pKa = 11.84VVSGDD25 pKa = 2.88TSEE28 pKa = 4.43RR29 pKa = 11.84VYY31 pKa = 10.55TAVLNYY37 pKa = 9.94CSTVRR42 pKa = 11.84QSYY45 pKa = 8.96QATTNVHH52 pKa = 4.73WRR54 pKa = 11.84LKK56 pKa = 10.14QRR58 pKa = 11.84GKK60 pKa = 10.34KK61 pKa = 9.54ASHH64 pKa = 6.32VSRR67 pKa = 11.84AHH69 pKa = 6.09AGISASFDD77 pKa = 3.38YY78 pKa = 11.21ADD80 pKa = 3.95NLKK83 pKa = 10.31QEE85 pKa = 3.89ILKK88 pKa = 8.93YY89 pKa = 10.22HH90 pKa = 5.97SRR92 pKa = 11.84HH93 pKa = 4.75VCTEE97 pKa = 3.67IVVGRR102 pKa = 11.84DD103 pKa = 3.05YY104 pKa = 11.42RR105 pKa = 11.84FDD107 pKa = 3.64FRR109 pKa = 11.84SLLHH113 pKa = 6.57HH114 pKa = 7.17AGACMAHH121 pKa = 5.9YY122 pKa = 10.34CLTGVLDD129 pKa = 4.64LNVLSSGKK137 pKa = 10.37SGGTRR142 pKa = 11.84VTVLSNADD150 pKa = 3.59GLSILATDD158 pKa = 4.1VLYY161 pKa = 11.01LPNALKK167 pKa = 10.69MMDD170 pKa = 3.52INIFNCIYY178 pKa = 10.57LLAAACEE185 pKa = 4.28CNIVFDD191 pKa = 5.26DD192 pKa = 4.66VILTGGTSAYY202 pKa = 10.04NVPALNSVTVEE213 pKa = 4.05YY214 pKa = 10.89APTLHH219 pKa = 6.71NYY221 pKa = 9.61ISVAFTSNGDD231 pKa = 3.76GGCDD235 pKa = 3.08GALALTAGLHH245 pKa = 4.72TVVTVVAHH253 pKa = 6.2SDD255 pKa = 2.97EE256 pKa = 4.69GGQMRR261 pKa = 11.84DD262 pKa = 3.15VLRR265 pKa = 11.84ALHH268 pKa = 5.69YY269 pKa = 10.66APAKK273 pKa = 9.81GIVVADD279 pKa = 3.94PNRR282 pKa = 11.84TVNHH286 pKa = 6.33SLPLLMYY293 pKa = 9.57EE294 pKa = 4.25PTWDD298 pKa = 4.13SVCGMWDD305 pKa = 3.46YY306 pKa = 11.61VAVATAGLVHH316 pKa = 6.94LCDD319 pKa = 3.83PCEE322 pKa = 4.49LIAHH326 pKa = 6.8DD327 pKa = 4.87VYY329 pKa = 10.36PTIITAPDD337 pKa = 3.56EE338 pKa = 4.45NNVRR342 pKa = 11.84AIEE345 pKa = 4.05ARR347 pKa = 11.84VEE349 pKa = 3.79AALPAFASIYY359 pKa = 9.47IHH361 pKa = 6.27NLAMFMGVGGDD372 pKa = 3.51DD373 pKa = 3.83CGRR376 pKa = 11.84AVDD379 pKa = 3.96TLVEE383 pKa = 4.27AGVYY387 pKa = 9.73IYY389 pKa = 11.2SLTGNDD395 pKa = 4.04ASRR398 pKa = 11.84HH399 pKa = 4.06MLKK402 pKa = 9.42GTMAPWFWVEE412 pKa = 3.84STGLFRR418 pKa = 11.84DD419 pKa = 4.37LTCFNIPGLSAGYY432 pKa = 9.78GPQAIYY438 pKa = 8.86GTAITQPAMQACEE451 pKa = 3.99YY452 pKa = 9.3TGTRR456 pKa = 11.84GSYY459 pKa = 10.39DD460 pKa = 3.7YY461 pKa = 11.7YY462 pKa = 10.96NVGWTSLRR470 pKa = 11.84KK471 pKa = 9.62HH472 pKa = 6.21PLLVLTNNRR481 pKa = 11.84AGDD484 pKa = 4.71GIAHH488 pKa = 6.04MEE490 pKa = 4.15VGRR493 pKa = 11.84DD494 pKa = 3.39AAIPWVLLGQPQRR507 pKa = 11.84RR508 pKa = 11.84EE509 pKa = 4.08CTAQGHH515 pKa = 5.22GTQTATCGHH524 pKa = 6.17TKK526 pKa = 10.21HH527 pKa = 6.51NRR529 pKa = 11.84GTLDD533 pKa = 3.02EE534 pKa = 4.84YY535 pKa = 10.28IWGRR539 pKa = 11.84FSTGLFHH546 pKa = 7.4PAEE549 pKa = 4.45LTTFTNVEE557 pKa = 4.15FRR559 pKa = 11.84VKK561 pKa = 10.24CWTEE565 pKa = 4.22DD566 pKa = 3.06NDD568 pKa = 4.49GNVLEE573 pKa = 4.91TGAPVKK579 pKa = 10.61DD580 pKa = 3.34IVEE583 pKa = 4.43GGVTVSVNAILLTNSTNHH601 pKa = 5.81IRR603 pKa = 11.84TVPAVRR609 pKa = 11.84RR610 pKa = 11.84SYY612 pKa = 10.7QAGAKK617 pKa = 9.14YY618 pKa = 10.54LEE620 pKa = 4.43EE621 pKa = 4.2ARR623 pKa = 11.84SRR625 pKa = 11.84GTMSTLNRR633 pKa = 11.84IIGGQLFRR641 pKa = 11.84DD642 pKa = 4.16LQPISKK648 pKa = 9.88RR649 pKa = 11.84GVPPQPEE656 pKa = 4.32PVCASMPVEE665 pKa = 4.19TTKK668 pKa = 10.68EE669 pKa = 3.97VSLSRR674 pKa = 11.84IGPIRR679 pKa = 11.84INQNVFRR686 pKa = 11.84PRR688 pKa = 11.84PQPTEE693 pKa = 3.97EE694 pKa = 4.23EE695 pKa = 4.19MHH697 pKa = 6.46EE698 pKa = 4.39EE699 pKa = 4.01PTPVALEE706 pKa = 4.05AEE708 pKa = 4.42PPGEE712 pKa = 4.11NVV714 pKa = 2.95
MM1 pKa = 7.58TSPIPQMNTSVFNLTRR17 pKa = 11.84GRR19 pKa = 11.84RR20 pKa = 11.84VVSGDD25 pKa = 2.88TSEE28 pKa = 4.43RR29 pKa = 11.84VYY31 pKa = 10.55TAVLNYY37 pKa = 9.94CSTVRR42 pKa = 11.84QSYY45 pKa = 8.96QATTNVHH52 pKa = 4.73WRR54 pKa = 11.84LKK56 pKa = 10.14QRR58 pKa = 11.84GKK60 pKa = 10.34KK61 pKa = 9.54ASHH64 pKa = 6.32VSRR67 pKa = 11.84AHH69 pKa = 6.09AGISASFDD77 pKa = 3.38YY78 pKa = 11.21ADD80 pKa = 3.95NLKK83 pKa = 10.31QEE85 pKa = 3.89ILKK88 pKa = 8.93YY89 pKa = 10.22HH90 pKa = 5.97SRR92 pKa = 11.84HH93 pKa = 4.75VCTEE97 pKa = 3.67IVVGRR102 pKa = 11.84DD103 pKa = 3.05YY104 pKa = 11.42RR105 pKa = 11.84FDD107 pKa = 3.64FRR109 pKa = 11.84SLLHH113 pKa = 6.57HH114 pKa = 7.17AGACMAHH121 pKa = 5.9YY122 pKa = 10.34CLTGVLDD129 pKa = 4.64LNVLSSGKK137 pKa = 10.37SGGTRR142 pKa = 11.84VTVLSNADD150 pKa = 3.59GLSILATDD158 pKa = 4.1VLYY161 pKa = 11.01LPNALKK167 pKa = 10.69MMDD170 pKa = 3.52INIFNCIYY178 pKa = 10.57LLAAACEE185 pKa = 4.28CNIVFDD191 pKa = 5.26DD192 pKa = 4.66VILTGGTSAYY202 pKa = 10.04NVPALNSVTVEE213 pKa = 4.05YY214 pKa = 10.89APTLHH219 pKa = 6.71NYY221 pKa = 9.61ISVAFTSNGDD231 pKa = 3.76GGCDD235 pKa = 3.08GALALTAGLHH245 pKa = 4.72TVVTVVAHH253 pKa = 6.2SDD255 pKa = 2.97EE256 pKa = 4.69GGQMRR261 pKa = 11.84DD262 pKa = 3.15VLRR265 pKa = 11.84ALHH268 pKa = 5.69YY269 pKa = 10.66APAKK273 pKa = 9.81GIVVADD279 pKa = 3.94PNRR282 pKa = 11.84TVNHH286 pKa = 6.33SLPLLMYY293 pKa = 9.57EE294 pKa = 4.25PTWDD298 pKa = 4.13SVCGMWDD305 pKa = 3.46YY306 pKa = 11.61VAVATAGLVHH316 pKa = 6.94LCDD319 pKa = 3.83PCEE322 pKa = 4.49LIAHH326 pKa = 6.8DD327 pKa = 4.87VYY329 pKa = 10.36PTIITAPDD337 pKa = 3.56EE338 pKa = 4.45NNVRR342 pKa = 11.84AIEE345 pKa = 4.05ARR347 pKa = 11.84VEE349 pKa = 3.79AALPAFASIYY359 pKa = 9.47IHH361 pKa = 6.27NLAMFMGVGGDD372 pKa = 3.51DD373 pKa = 3.83CGRR376 pKa = 11.84AVDD379 pKa = 3.96TLVEE383 pKa = 4.27AGVYY387 pKa = 9.73IYY389 pKa = 11.2SLTGNDD395 pKa = 4.04ASRR398 pKa = 11.84HH399 pKa = 4.06MLKK402 pKa = 9.42GTMAPWFWVEE412 pKa = 3.84STGLFRR418 pKa = 11.84DD419 pKa = 4.37LTCFNIPGLSAGYY432 pKa = 9.78GPQAIYY438 pKa = 8.86GTAITQPAMQACEE451 pKa = 3.99YY452 pKa = 9.3TGTRR456 pKa = 11.84GSYY459 pKa = 10.39DD460 pKa = 3.7YY461 pKa = 11.7YY462 pKa = 10.96NVGWTSLRR470 pKa = 11.84KK471 pKa = 9.62HH472 pKa = 6.21PLLVLTNNRR481 pKa = 11.84AGDD484 pKa = 4.71GIAHH488 pKa = 6.04MEE490 pKa = 4.15VGRR493 pKa = 11.84DD494 pKa = 3.39AAIPWVLLGQPQRR507 pKa = 11.84RR508 pKa = 11.84EE509 pKa = 4.08CTAQGHH515 pKa = 5.22GTQTATCGHH524 pKa = 6.17TKK526 pKa = 10.21HH527 pKa = 6.51NRR529 pKa = 11.84GTLDD533 pKa = 3.02EE534 pKa = 4.84YY535 pKa = 10.28IWGRR539 pKa = 11.84FSTGLFHH546 pKa = 7.4PAEE549 pKa = 4.45LTTFTNVEE557 pKa = 4.15FRR559 pKa = 11.84VKK561 pKa = 10.24CWTEE565 pKa = 4.22DD566 pKa = 3.06NDD568 pKa = 4.49GNVLEE573 pKa = 4.91TGAPVKK579 pKa = 10.61DD580 pKa = 3.34IVEE583 pKa = 4.43GGVTVSVNAILLTNSTNHH601 pKa = 5.81IRR603 pKa = 11.84TVPAVRR609 pKa = 11.84RR610 pKa = 11.84SYY612 pKa = 10.7QAGAKK617 pKa = 9.14YY618 pKa = 10.54LEE620 pKa = 4.43EE621 pKa = 4.2ARR623 pKa = 11.84SRR625 pKa = 11.84GTMSTLNRR633 pKa = 11.84IIGGQLFRR641 pKa = 11.84DD642 pKa = 4.16LQPISKK648 pKa = 9.88RR649 pKa = 11.84GVPPQPEE656 pKa = 4.32PVCASMPVEE665 pKa = 4.19TTKK668 pKa = 10.68EE669 pKa = 3.97VSLSRR674 pKa = 11.84IGPIRR679 pKa = 11.84INQNVFRR686 pKa = 11.84PRR688 pKa = 11.84PQPTEE693 pKa = 3.97EE694 pKa = 4.23EE695 pKa = 4.19MHH697 pKa = 6.46EE698 pKa = 4.39EE699 pKa = 4.01PTPVALEE706 pKa = 4.05AEE708 pKa = 4.42PPGEE712 pKa = 4.11NVV714 pKa = 2.95
Molecular weight: 77.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q83072|Q83072_9VIRU Major capsid protein OS=Leishmania RNA virus 2 - 1 OX=39116 PE=4 SV=1
MM1 pKa = 7.02VCRR4 pKa = 11.84SVRR7 pKa = 11.84PAILILTPVDD17 pKa = 3.21AGKK20 pKa = 10.1NRR22 pKa = 11.84VMGWHH27 pKa = 6.48PAPYY31 pKa = 9.58VFQSLVYY38 pKa = 9.56YY39 pKa = 10.78
MM1 pKa = 7.02VCRR4 pKa = 11.84SVRR7 pKa = 11.84PAILILTPVDD17 pKa = 3.21AGKK20 pKa = 10.1NRR22 pKa = 11.84VMGWHH27 pKa = 6.48PAPYY31 pKa = 9.58VFQSLVYY38 pKa = 9.56YY39 pKa = 10.78
Molecular weight: 4.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1530 |
39 |
777 |
510.0 |
56.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.954 ± 0.199 | 2.484 ± 0.029 |
5.033 ± 0.414 | 5.359 ± 0.857 |
2.549 ± 0.019 | 6.797 ± 0.994 |
3.791 ± 0.275 | 4.575 ± 0.155 |
4.052 ± 1.259 | 8.954 ± 0.516 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.745 ± 0.402 | 4.248 ± 0.566 |
4.314 ± 1.081 | 2.418 ± 0.167 |
6.405 ± 0.382 | 6.34 ± 0.454 |
6.144 ± 1.471 | 8.954 ± 0.921 |
1.438 ± 0.192 | 4.444 ± 0.558 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
