
Marichromatium purpuratum 984
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae; Marichromatium; Marichromatium purpuratum
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
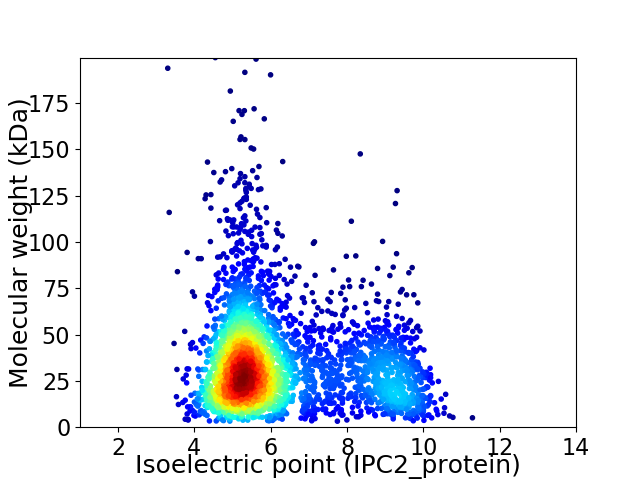
Virtual 2D-PAGE plot for 3249 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W0E7P4|W0E7P4_MARPU Uncharacterized protein OS=Marichromatium purpuratum 984 OX=765910 GN=MARPU_10805 PE=4 SV=1
MM1 pKa = 7.75DD2 pKa = 3.93VSRR5 pKa = 11.84LTLCSLLVAAALAPHH20 pKa = 7.08AASADD25 pKa = 3.14DD26 pKa = 3.71WQRR29 pKa = 11.84IDD31 pKa = 3.24AAAPFVDD38 pKa = 3.23ADD40 pKa = 3.94GVFRR44 pKa = 11.84TPACSGSPRR53 pKa = 11.84LEE55 pKa = 4.19EE56 pKa = 4.58GEE58 pKa = 4.6VVATDD63 pKa = 2.95TDD65 pKa = 3.95YY66 pKa = 11.97AFFVRR71 pKa = 11.84DD72 pKa = 3.58GDD74 pKa = 4.01PEE76 pKa = 4.17RR77 pKa = 11.84LLLFFDD83 pKa = 4.89GGGACWDD90 pKa = 3.76AGTCIDD96 pKa = 4.7AALDD100 pKa = 3.76GNVLYY105 pKa = 10.39FPEE108 pKa = 4.21VDD110 pKa = 3.46EE111 pKa = 5.27TPDD114 pKa = 3.22EE115 pKa = 4.16LAGFGGVFDD124 pKa = 6.42LDD126 pKa = 3.61RR127 pKa = 11.84TDD129 pKa = 3.72NPLRR133 pKa = 11.84DD134 pKa = 3.83HH135 pKa = 6.39LQVYY139 pKa = 9.74LPYY142 pKa = 9.94CTGDD146 pKa = 3.31LHH148 pKa = 8.36LGARR152 pKa = 11.84DD153 pKa = 3.51TSYY156 pKa = 10.97PLPDD160 pKa = 4.12GGDD163 pKa = 2.85WTIYY167 pKa = 10.27HH168 pKa = 7.09RR169 pKa = 11.84GHH171 pKa = 7.16DD172 pKa = 3.63NVIAVLEE179 pKa = 4.04YY180 pKa = 10.86LADD183 pKa = 3.74YY184 pKa = 9.25YY185 pKa = 11.46ANEE188 pKa = 4.17VGEE191 pKa = 4.21PPEE194 pKa = 4.51EE195 pKa = 3.95IVLVGASAGGYY206 pKa = 8.7GVLYY210 pKa = 10.06NYY212 pKa = 7.96PALARR217 pKa = 11.84RR218 pKa = 11.84FPGVEE223 pKa = 4.11RR224 pKa = 11.84IRR226 pKa = 11.84VLVDD230 pKa = 2.9SSNGVISEE238 pKa = 4.32DD239 pKa = 3.26LFEE242 pKa = 4.96RR243 pKa = 11.84ALEE246 pKa = 4.17ADD248 pKa = 4.42GVWGAQEE255 pKa = 4.17NLDD258 pKa = 4.19PEE260 pKa = 4.32IASAFDD266 pKa = 4.01DD267 pKa = 4.58GAGQLVPGLFNTLGWQYY284 pKa = 10.78PDD286 pKa = 3.77ARR288 pKa = 11.84FGQYY292 pKa = 7.83TRR294 pKa = 11.84AYY296 pKa = 10.3DD297 pKa = 3.53AVQVLFFNVSSNLGNPEE314 pKa = 3.56RR315 pKa = 11.84WFDD318 pKa = 3.7PVYY321 pKa = 11.03LLLSGLEE328 pKa = 3.78WTTRR332 pKa = 11.84ARR334 pKa = 11.84ASMWGTAFTTWNYY347 pKa = 10.55RR348 pKa = 11.84FYY350 pKa = 10.93LAQGQTHH357 pKa = 5.58MVSFYY362 pKa = 11.34DD363 pKa = 4.34DD364 pKa = 4.16FFNEE368 pKa = 4.21DD369 pKa = 3.19SAGGIEE375 pKa = 4.05LSDD378 pKa = 3.33WFGDD382 pKa = 3.59MIEE385 pKa = 4.28RR386 pKa = 11.84RR387 pKa = 11.84WTFGSDD393 pKa = 2.33WRR395 pKa = 11.84NASCVPACLPPEE407 pKa = 4.25GLDD410 pKa = 4.39LSTLL414 pKa = 3.72
MM1 pKa = 7.75DD2 pKa = 3.93VSRR5 pKa = 11.84LTLCSLLVAAALAPHH20 pKa = 7.08AASADD25 pKa = 3.14DD26 pKa = 3.71WQRR29 pKa = 11.84IDD31 pKa = 3.24AAAPFVDD38 pKa = 3.23ADD40 pKa = 3.94GVFRR44 pKa = 11.84TPACSGSPRR53 pKa = 11.84LEE55 pKa = 4.19EE56 pKa = 4.58GEE58 pKa = 4.6VVATDD63 pKa = 2.95TDD65 pKa = 3.95YY66 pKa = 11.97AFFVRR71 pKa = 11.84DD72 pKa = 3.58GDD74 pKa = 4.01PEE76 pKa = 4.17RR77 pKa = 11.84LLLFFDD83 pKa = 4.89GGGACWDD90 pKa = 3.76AGTCIDD96 pKa = 4.7AALDD100 pKa = 3.76GNVLYY105 pKa = 10.39FPEE108 pKa = 4.21VDD110 pKa = 3.46EE111 pKa = 5.27TPDD114 pKa = 3.22EE115 pKa = 4.16LAGFGGVFDD124 pKa = 6.42LDD126 pKa = 3.61RR127 pKa = 11.84TDD129 pKa = 3.72NPLRR133 pKa = 11.84DD134 pKa = 3.83HH135 pKa = 6.39LQVYY139 pKa = 9.74LPYY142 pKa = 9.94CTGDD146 pKa = 3.31LHH148 pKa = 8.36LGARR152 pKa = 11.84DD153 pKa = 3.51TSYY156 pKa = 10.97PLPDD160 pKa = 4.12GGDD163 pKa = 2.85WTIYY167 pKa = 10.27HH168 pKa = 7.09RR169 pKa = 11.84GHH171 pKa = 7.16DD172 pKa = 3.63NVIAVLEE179 pKa = 4.04YY180 pKa = 10.86LADD183 pKa = 3.74YY184 pKa = 9.25YY185 pKa = 11.46ANEE188 pKa = 4.17VGEE191 pKa = 4.21PPEE194 pKa = 4.51EE195 pKa = 3.95IVLVGASAGGYY206 pKa = 8.7GVLYY210 pKa = 10.06NYY212 pKa = 7.96PALARR217 pKa = 11.84RR218 pKa = 11.84FPGVEE223 pKa = 4.11RR224 pKa = 11.84IRR226 pKa = 11.84VLVDD230 pKa = 2.9SSNGVISEE238 pKa = 4.32DD239 pKa = 3.26LFEE242 pKa = 4.96RR243 pKa = 11.84ALEE246 pKa = 4.17ADD248 pKa = 4.42GVWGAQEE255 pKa = 4.17NLDD258 pKa = 4.19PEE260 pKa = 4.32IASAFDD266 pKa = 4.01DD267 pKa = 4.58GAGQLVPGLFNTLGWQYY284 pKa = 10.78PDD286 pKa = 3.77ARR288 pKa = 11.84FGQYY292 pKa = 7.83TRR294 pKa = 11.84AYY296 pKa = 10.3DD297 pKa = 3.53AVQVLFFNVSSNLGNPEE314 pKa = 3.56RR315 pKa = 11.84WFDD318 pKa = 3.7PVYY321 pKa = 11.03LLLSGLEE328 pKa = 3.78WTTRR332 pKa = 11.84ARR334 pKa = 11.84ASMWGTAFTTWNYY347 pKa = 10.55RR348 pKa = 11.84FYY350 pKa = 10.93LAQGQTHH357 pKa = 5.58MVSFYY362 pKa = 11.34DD363 pKa = 4.34DD364 pKa = 4.16FFNEE368 pKa = 4.21DD369 pKa = 3.19SAGGIEE375 pKa = 4.05LSDD378 pKa = 3.33WFGDD382 pKa = 3.59MIEE385 pKa = 4.28RR386 pKa = 11.84RR387 pKa = 11.84WTFGSDD393 pKa = 2.33WRR395 pKa = 11.84NASCVPACLPPEE407 pKa = 4.25GLDD410 pKa = 4.39LSTLL414 pKa = 3.72
Molecular weight: 45.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W0E3D9|W0E3D9_MARPU Diguanylate cyclase OS=Marichromatium purpuratum 984 OX=765910 GN=MARPU_07445 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.48RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84SATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.2VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LALL44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.48RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84SATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.2VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.29GRR39 pKa = 11.84ARR41 pKa = 11.84LALL44 pKa = 3.93
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1091096 |
30 |
1974 |
335.8 |
36.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.579 ± 0.06 | 1.023 ± 0.016 |
5.779 ± 0.035 | 6.605 ± 0.044 |
3.164 ± 0.027 | 8.258 ± 0.038 |
2.384 ± 0.026 | 4.439 ± 0.03 |
1.926 ± 0.027 | 11.958 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.042 ± 0.019 | 2.057 ± 0.023 |
5.368 ± 0.037 | 3.451 ± 0.03 |
8.681 ± 0.052 | 4.611 ± 0.032 |
4.889 ± 0.033 | 7.197 ± 0.036 |
1.391 ± 0.02 | 2.196 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
