
Rosellinia necatrix megabirnavirus 1 (isolate -/Japan/W779/2001) (RnMBV1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Megabirnaviridae; Megabirnavirus; Rosellinia necatrix megabirnavirus 1
Average proteome isoelectric point is 5.73
Get precalculated fractions of proteins
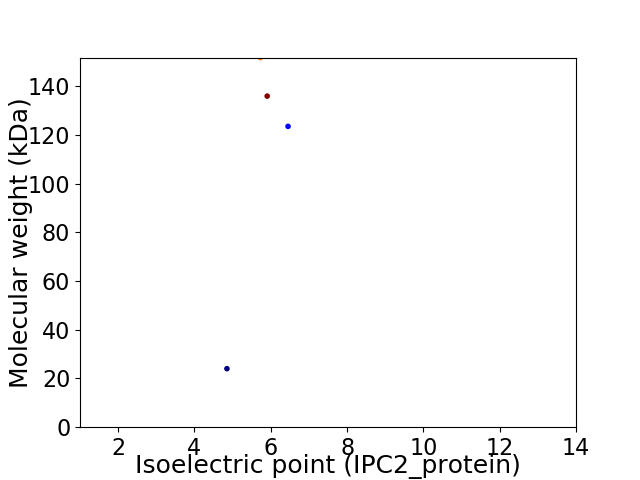
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D0FZL3|D0FZL3_RMBV1 Uncharacterized protein OS=Rosellinia necatrix megabirnavirus 1 (isolate -/Japan/W779/2001) OX=658904 PE=4 SV=1
MM1 pKa = 7.72LARR4 pKa = 11.84ALCSDD9 pKa = 4.31RR10 pKa = 11.84ALACLKK16 pKa = 10.34SLRR19 pKa = 11.84AGNVARR25 pKa = 11.84IRR27 pKa = 11.84RR28 pKa = 11.84EE29 pKa = 3.24IAVVVKK35 pKa = 9.5VVKK38 pKa = 10.26CCRR41 pKa = 11.84LLVEE45 pKa = 4.43RR46 pKa = 11.84VARR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 9.98VSEE54 pKa = 4.1GVPSGSTDD62 pKa = 3.47VVPLEE67 pKa = 4.12VSGLTEE73 pKa = 3.71ATVFDD78 pKa = 4.52HH79 pKa = 7.46LLLQLVQEE87 pKa = 4.59TLLPSPVSGGAVGVTHH103 pKa = 6.47RR104 pKa = 11.84TAEE107 pKa = 3.99EE108 pKa = 3.8AAPASALQVGTSAGTDD124 pKa = 3.32VIPDD128 pKa = 3.51VVLTSVVSYY137 pKa = 10.36HH138 pKa = 6.56RR139 pKa = 11.84EE140 pKa = 3.8AVEE143 pKa = 4.44HH144 pKa = 6.34IFVAAEE150 pKa = 3.59ARR152 pKa = 11.84AGTRR156 pKa = 11.84EE157 pKa = 4.01VQLLLASVDD166 pKa = 3.63EE167 pKa = 4.46SLVRR171 pKa = 11.84AVYY174 pKa = 8.91PAGLTDD180 pKa = 4.3PVLTLLAHH188 pKa = 7.38DD189 pKa = 4.88YY190 pKa = 11.12ADD192 pKa = 3.96SDD194 pKa = 3.26IGQPFTLHH202 pKa = 6.21YY203 pKa = 10.81SEE205 pKa = 6.16LDD207 pKa = 3.63LGCLSSLTPEE217 pKa = 4.59EE218 pKa = 4.21EE219 pKa = 4.19TGDD222 pKa = 4.21ADD224 pKa = 5.12DD225 pKa = 5.94GYY227 pKa = 12.01
MM1 pKa = 7.72LARR4 pKa = 11.84ALCSDD9 pKa = 4.31RR10 pKa = 11.84ALACLKK16 pKa = 10.34SLRR19 pKa = 11.84AGNVARR25 pKa = 11.84IRR27 pKa = 11.84RR28 pKa = 11.84EE29 pKa = 3.24IAVVVKK35 pKa = 9.5VVKK38 pKa = 10.26CCRR41 pKa = 11.84LLVEE45 pKa = 4.43RR46 pKa = 11.84VARR49 pKa = 11.84RR50 pKa = 11.84KK51 pKa = 9.98VSEE54 pKa = 4.1GVPSGSTDD62 pKa = 3.47VVPLEE67 pKa = 4.12VSGLTEE73 pKa = 3.71ATVFDD78 pKa = 4.52HH79 pKa = 7.46LLLQLVQEE87 pKa = 4.59TLLPSPVSGGAVGVTHH103 pKa = 6.47RR104 pKa = 11.84TAEE107 pKa = 3.99EE108 pKa = 3.8AAPASALQVGTSAGTDD124 pKa = 3.32VIPDD128 pKa = 3.51VVLTSVVSYY137 pKa = 10.36HH138 pKa = 6.56RR139 pKa = 11.84EE140 pKa = 3.8AVEE143 pKa = 4.44HH144 pKa = 6.34IFVAAEE150 pKa = 3.59ARR152 pKa = 11.84AGTRR156 pKa = 11.84EE157 pKa = 4.01VQLLLASVDD166 pKa = 3.63EE167 pKa = 4.46SLVRR171 pKa = 11.84AVYY174 pKa = 8.91PAGLTDD180 pKa = 4.3PVLTLLAHH188 pKa = 7.38DD189 pKa = 4.88YY190 pKa = 11.12ADD192 pKa = 3.96SDD194 pKa = 3.26IGQPFTLHH202 pKa = 6.21YY203 pKa = 10.81SEE205 pKa = 6.16LDD207 pKa = 3.63LGCLSSLTPEE217 pKa = 4.59EE218 pKa = 4.21EE219 pKa = 4.19TGDD222 pKa = 4.21ADD224 pKa = 5.12DD225 pKa = 5.94GYY227 pKa = 12.01
Molecular weight: 24.03 kDa
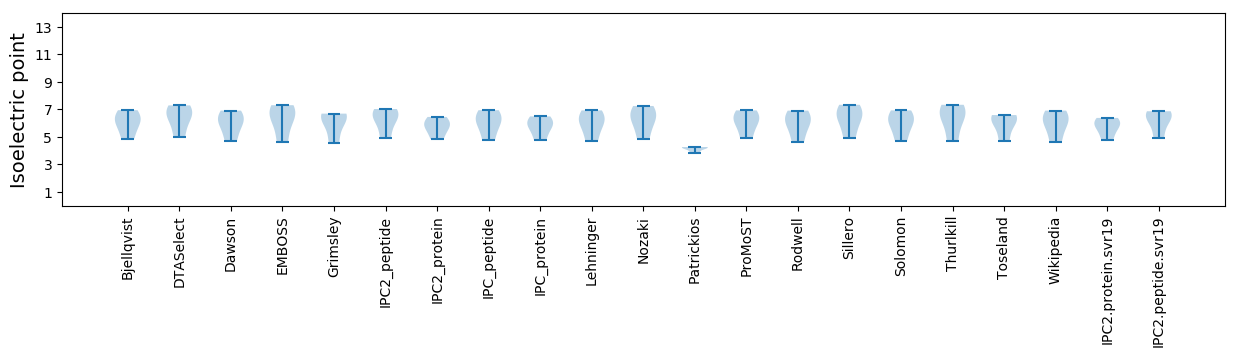
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0FZL2|D0FZL2_RMBV1 Uncharacterized protein OS=Rosellinia necatrix megabirnavirus 1 (isolate -/Japan/W779/2001) OX=658904 PE=4 SV=1
MM1 pKa = 8.17DD2 pKa = 4.87GRR4 pKa = 11.84HH5 pKa = 6.37DD6 pKa = 3.75PDD8 pKa = 3.71AALFAQDD15 pKa = 3.36NVYY18 pKa = 10.1FSSVADD24 pKa = 3.5VEE26 pKa = 4.28RR27 pKa = 11.84ATWSDD32 pKa = 2.8IVAKK36 pKa = 10.58ARR38 pKa = 11.84ADD40 pKa = 3.16GWQGRR45 pKa = 11.84DD46 pKa = 3.24VLQEE50 pKa = 3.89FGGGQTLARR59 pKa = 11.84LVRR62 pKa = 11.84NSAIPDD68 pKa = 3.52DD69 pKa = 4.05FRR71 pKa = 11.84NSIVTAKK78 pKa = 9.88GAEE81 pKa = 3.91WLLLRR86 pKa = 11.84SVAHH90 pKa = 5.03VALRR94 pKa = 11.84LSHH97 pKa = 6.75NSYY100 pKa = 11.02KK101 pKa = 10.45KK102 pKa = 10.7LKK104 pKa = 9.87TSILTKK110 pKa = 10.22RR111 pKa = 11.84GCNGSRR117 pKa = 11.84RR118 pKa = 11.84LQAVRR123 pKa = 11.84SVMEE127 pKa = 3.8AAFAGNVPDD136 pKa = 4.12WLAEE140 pKa = 3.84MEE142 pKa = 4.12MLEE145 pKa = 4.29RR146 pKa = 11.84CGPEE150 pKa = 3.62YY151 pKa = 10.86AAVTRR156 pKa = 11.84IVGDD160 pKa = 4.26DD161 pKa = 3.57EE162 pKa = 4.96FDD164 pKa = 3.85PLGTISAVAACEE176 pKa = 4.03GGLNLFVFDD185 pKa = 5.02GPAEE189 pKa = 4.44GGSGHH194 pKa = 5.96EE195 pKa = 4.05AVVVEE200 pKa = 4.51GDD202 pKa = 3.37PGEE205 pKa = 4.3AGAYY209 pKa = 8.03VEE211 pKa = 5.52RR212 pKa = 11.84RR213 pKa = 11.84LKK215 pKa = 10.8LEE217 pKa = 3.84KK218 pKa = 10.16RR219 pKa = 11.84SLAKK223 pKa = 10.47GVINVEE229 pKa = 4.03LKK231 pKa = 10.73KK232 pKa = 11.05VITSSFLLSCEE243 pKa = 4.39VRR245 pKa = 11.84DD246 pKa = 3.72LDD248 pKa = 4.45YY249 pKa = 11.44AKK251 pKa = 10.41HH252 pKa = 6.24CLRR255 pKa = 11.84ILFPITDD262 pKa = 3.47MNLYY266 pKa = 10.3LRR268 pKa = 11.84ARR270 pKa = 11.84LNYY273 pKa = 8.55GCLIRR278 pKa = 11.84VIFKK282 pKa = 10.96LLDD285 pKa = 3.58DD286 pKa = 5.83RR287 pKa = 11.84ILDD290 pKa = 3.99LSHH293 pKa = 7.11LSSVCTNAAGLGHH306 pKa = 6.12QAFACLILTLASGAEE321 pKa = 3.76RR322 pKa = 11.84RR323 pKa = 11.84NCAVVMLQQGIYY335 pKa = 9.0RR336 pKa = 11.84GGMKK340 pKa = 10.11SVEE343 pKa = 3.99KK344 pKa = 10.18RR345 pKa = 11.84LKK347 pKa = 9.98EE348 pKa = 3.43IHH350 pKa = 5.27TAARR354 pKa = 11.84RR355 pKa = 11.84ARR357 pKa = 11.84WLPSYY362 pKa = 10.5LAGVCTDD369 pKa = 4.25PSVMLYY375 pKa = 10.3WNIALGRR382 pKa = 11.84FYY384 pKa = 11.2LDD386 pKa = 3.15YY387 pKa = 10.88AANDD391 pKa = 3.65DD392 pKa = 4.14TVKK395 pKa = 10.93DD396 pKa = 3.25RR397 pKa = 11.84TMYY400 pKa = 9.76MGEE403 pKa = 3.92HH404 pKa = 5.59VAYY407 pKa = 10.14DD408 pKa = 3.51AASRR412 pKa = 11.84MYY414 pKa = 10.69TSDD417 pKa = 2.71QHH419 pKa = 5.89AANMQRR425 pKa = 11.84FFNARR430 pKa = 11.84KK431 pKa = 9.41QDD433 pKa = 3.56AQKK436 pKa = 9.74VWSSSEE442 pKa = 3.84VDD444 pKa = 4.13TLDD447 pKa = 3.16SWLLRR452 pKa = 11.84YY453 pKa = 9.38VQFGSSGSAGGHH465 pKa = 5.98SGDD468 pKa = 4.91EE469 pKa = 3.92FDD471 pKa = 5.08IDD473 pKa = 3.69HH474 pKa = 6.74AVSKK478 pKa = 10.53RR479 pKa = 11.84LWLSNRR485 pKa = 11.84DD486 pKa = 3.41TEE488 pKa = 4.48TIAWYY493 pKa = 9.92VLKK496 pKa = 10.67QPAGASTVTIVKK508 pKa = 10.24RR509 pKa = 11.84EE510 pKa = 4.02AGKK513 pKa = 10.17LRR515 pKa = 11.84QLLPGRR521 pKa = 11.84IPHH524 pKa = 6.95WIVEE528 pKa = 4.24SLLINEE534 pKa = 4.68IEE536 pKa = 4.25SAIYY540 pKa = 10.17RR541 pKa = 11.84SMPMEE546 pKa = 4.98LEE548 pKa = 4.31LDD550 pKa = 3.65AQSEE554 pKa = 4.76YY555 pKa = 10.21TSVMEE560 pKa = 4.0RR561 pKa = 11.84RR562 pKa = 11.84RR563 pKa = 11.84RR564 pKa = 11.84MMLGEE569 pKa = 4.24SVACVDD575 pKa = 3.2WADD578 pKa = 3.9FNITHH583 pKa = 6.27TLKK586 pKa = 11.12DD587 pKa = 3.26MSEE590 pKa = 4.2YY591 pKa = 10.64FLALGEE597 pKa = 4.13AAQRR601 pKa = 11.84ACVEE605 pKa = 4.39NNFVTYY611 pKa = 10.73GGVNYY616 pKa = 10.58GKK618 pKa = 11.15GEE620 pKa = 3.9FLSLCANWCSKK631 pKa = 10.93ALYY634 pKa = 10.62NMWLKK639 pKa = 11.13NGADD643 pKa = 3.51KK644 pKa = 11.3DD645 pKa = 3.89SDD647 pKa = 4.2YY648 pKa = 10.95EE649 pKa = 3.95AHH651 pKa = 7.41ARR653 pKa = 11.84GLWSGWRR660 pKa = 11.84TTSFINCSFNVAYY673 pKa = 10.49CEE675 pKa = 4.61SNFDD679 pKa = 3.91SMHH682 pKa = 6.86HH683 pKa = 6.39IFNSDD688 pKa = 2.96MPLYY692 pKa = 9.86RR693 pKa = 11.84VHH695 pKa = 8.05AGDD698 pKa = 5.47DD699 pKa = 3.67FFGTYY704 pKa = 10.25AEE706 pKa = 5.31EE707 pKa = 3.98IDD709 pKa = 3.3AMRR712 pKa = 11.84FVSSMPLAKK721 pKa = 10.35HH722 pKa = 5.41EE723 pKa = 4.43VNAQKK728 pKa = 10.63QLVGGEE734 pKa = 3.76MGEE737 pKa = 4.3FLRR740 pKa = 11.84HH741 pKa = 5.39EE742 pKa = 4.43YY743 pKa = 10.6FRR745 pKa = 11.84DD746 pKa = 3.47GTVRR750 pKa = 11.84GSVMRR755 pKa = 11.84SIGSFVGSDD764 pKa = 3.29LQAPQLYY771 pKa = 10.37SGVAQCQGANEE782 pKa = 4.96AINLLIRR789 pKa = 11.84RR790 pKa = 11.84GFDD793 pKa = 2.95LRR795 pKa = 11.84AAEE798 pKa = 4.43SIRR801 pKa = 11.84YY802 pKa = 7.67TIVQYY807 pKa = 10.19WGTVKK812 pKa = 10.63EE813 pKa = 4.51PGGSSATPSVKK824 pKa = 10.3LLALSAHH831 pKa = 6.35SGGLGCPRR839 pKa = 11.84FGEE842 pKa = 4.26LATVTRR848 pKa = 11.84EE849 pKa = 3.88RR850 pKa = 11.84LHH852 pKa = 6.83GIPVMRR858 pKa = 11.84DD859 pKa = 3.33VLSKK863 pKa = 10.77LAAPRR868 pKa = 11.84NMEE871 pKa = 4.17RR872 pKa = 11.84LLHH875 pKa = 6.62AFQAKK880 pKa = 8.79IRR882 pKa = 11.84RR883 pKa = 11.84FGYY886 pKa = 9.79IEE888 pKa = 4.06EE889 pKa = 4.87GFTALRR895 pKa = 11.84NAAYY899 pKa = 9.97NATYY903 pKa = 10.52GSDD906 pKa = 3.52LPRR909 pKa = 11.84EE910 pKa = 4.29IKK912 pKa = 10.84LEE914 pKa = 4.05ADD916 pKa = 3.43GEE918 pKa = 4.22LRR920 pKa = 11.84RR921 pKa = 11.84AQVAWINEE929 pKa = 4.29CNLKK933 pKa = 10.0VPEE936 pKa = 4.51MTVAWYY942 pKa = 8.88PSVPSKK948 pKa = 10.69VLEE951 pKa = 3.94YY952 pKa = 10.9LEE954 pKa = 5.17RR955 pKa = 11.84LIACAEE961 pKa = 4.08KK962 pKa = 10.68GLEE965 pKa = 4.19PEE967 pKa = 4.51YY968 pKa = 10.62PMHH971 pKa = 7.34DD972 pKa = 4.01LNGMAVAARR981 pKa = 11.84ATALGAFSALDD992 pKa = 3.23SGLNFLGDD1000 pKa = 3.34EE1001 pKa = 4.52AGRR1004 pKa = 11.84VAGFDD1009 pKa = 3.24AVVAMSRR1016 pKa = 11.84GRR1018 pKa = 11.84AGTILNRR1025 pKa = 11.84MRR1027 pKa = 11.84LYY1029 pKa = 10.75LPEE1032 pKa = 4.8IAVRR1036 pKa = 11.84ALVEE1040 pKa = 4.2HH1041 pKa = 7.04RR1042 pKa = 11.84WNVPTSTGGVVPAQLRR1058 pKa = 11.84GIVFSGLNYY1067 pKa = 10.49ALWYY1071 pKa = 9.23AKK1073 pKa = 10.23NWMPVNAGSLDD1084 pKa = 3.55WLTSYY1089 pKa = 11.14CDD1091 pKa = 3.3AVNIRR1096 pKa = 11.84IANLYY1101 pKa = 10.01KK1102 pKa = 10.64SSLSKK1107 pKa = 9.75MMKK1110 pKa = 8.62MM1111 pKa = 4.54
MM1 pKa = 8.17DD2 pKa = 4.87GRR4 pKa = 11.84HH5 pKa = 6.37DD6 pKa = 3.75PDD8 pKa = 3.71AALFAQDD15 pKa = 3.36NVYY18 pKa = 10.1FSSVADD24 pKa = 3.5VEE26 pKa = 4.28RR27 pKa = 11.84ATWSDD32 pKa = 2.8IVAKK36 pKa = 10.58ARR38 pKa = 11.84ADD40 pKa = 3.16GWQGRR45 pKa = 11.84DD46 pKa = 3.24VLQEE50 pKa = 3.89FGGGQTLARR59 pKa = 11.84LVRR62 pKa = 11.84NSAIPDD68 pKa = 3.52DD69 pKa = 4.05FRR71 pKa = 11.84NSIVTAKK78 pKa = 9.88GAEE81 pKa = 3.91WLLLRR86 pKa = 11.84SVAHH90 pKa = 5.03VALRR94 pKa = 11.84LSHH97 pKa = 6.75NSYY100 pKa = 11.02KK101 pKa = 10.45KK102 pKa = 10.7LKK104 pKa = 9.87TSILTKK110 pKa = 10.22RR111 pKa = 11.84GCNGSRR117 pKa = 11.84RR118 pKa = 11.84LQAVRR123 pKa = 11.84SVMEE127 pKa = 3.8AAFAGNVPDD136 pKa = 4.12WLAEE140 pKa = 3.84MEE142 pKa = 4.12MLEE145 pKa = 4.29RR146 pKa = 11.84CGPEE150 pKa = 3.62YY151 pKa = 10.86AAVTRR156 pKa = 11.84IVGDD160 pKa = 4.26DD161 pKa = 3.57EE162 pKa = 4.96FDD164 pKa = 3.85PLGTISAVAACEE176 pKa = 4.03GGLNLFVFDD185 pKa = 5.02GPAEE189 pKa = 4.44GGSGHH194 pKa = 5.96EE195 pKa = 4.05AVVVEE200 pKa = 4.51GDD202 pKa = 3.37PGEE205 pKa = 4.3AGAYY209 pKa = 8.03VEE211 pKa = 5.52RR212 pKa = 11.84RR213 pKa = 11.84LKK215 pKa = 10.8LEE217 pKa = 3.84KK218 pKa = 10.16RR219 pKa = 11.84SLAKK223 pKa = 10.47GVINVEE229 pKa = 4.03LKK231 pKa = 10.73KK232 pKa = 11.05VITSSFLLSCEE243 pKa = 4.39VRR245 pKa = 11.84DD246 pKa = 3.72LDD248 pKa = 4.45YY249 pKa = 11.44AKK251 pKa = 10.41HH252 pKa = 6.24CLRR255 pKa = 11.84ILFPITDD262 pKa = 3.47MNLYY266 pKa = 10.3LRR268 pKa = 11.84ARR270 pKa = 11.84LNYY273 pKa = 8.55GCLIRR278 pKa = 11.84VIFKK282 pKa = 10.96LLDD285 pKa = 3.58DD286 pKa = 5.83RR287 pKa = 11.84ILDD290 pKa = 3.99LSHH293 pKa = 7.11LSSVCTNAAGLGHH306 pKa = 6.12QAFACLILTLASGAEE321 pKa = 3.76RR322 pKa = 11.84RR323 pKa = 11.84NCAVVMLQQGIYY335 pKa = 9.0RR336 pKa = 11.84GGMKK340 pKa = 10.11SVEE343 pKa = 3.99KK344 pKa = 10.18RR345 pKa = 11.84LKK347 pKa = 9.98EE348 pKa = 3.43IHH350 pKa = 5.27TAARR354 pKa = 11.84RR355 pKa = 11.84ARR357 pKa = 11.84WLPSYY362 pKa = 10.5LAGVCTDD369 pKa = 4.25PSVMLYY375 pKa = 10.3WNIALGRR382 pKa = 11.84FYY384 pKa = 11.2LDD386 pKa = 3.15YY387 pKa = 10.88AANDD391 pKa = 3.65DD392 pKa = 4.14TVKK395 pKa = 10.93DD396 pKa = 3.25RR397 pKa = 11.84TMYY400 pKa = 9.76MGEE403 pKa = 3.92HH404 pKa = 5.59VAYY407 pKa = 10.14DD408 pKa = 3.51AASRR412 pKa = 11.84MYY414 pKa = 10.69TSDD417 pKa = 2.71QHH419 pKa = 5.89AANMQRR425 pKa = 11.84FFNARR430 pKa = 11.84KK431 pKa = 9.41QDD433 pKa = 3.56AQKK436 pKa = 9.74VWSSSEE442 pKa = 3.84VDD444 pKa = 4.13TLDD447 pKa = 3.16SWLLRR452 pKa = 11.84YY453 pKa = 9.38VQFGSSGSAGGHH465 pKa = 5.98SGDD468 pKa = 4.91EE469 pKa = 3.92FDD471 pKa = 5.08IDD473 pKa = 3.69HH474 pKa = 6.74AVSKK478 pKa = 10.53RR479 pKa = 11.84LWLSNRR485 pKa = 11.84DD486 pKa = 3.41TEE488 pKa = 4.48TIAWYY493 pKa = 9.92VLKK496 pKa = 10.67QPAGASTVTIVKK508 pKa = 10.24RR509 pKa = 11.84EE510 pKa = 4.02AGKK513 pKa = 10.17LRR515 pKa = 11.84QLLPGRR521 pKa = 11.84IPHH524 pKa = 6.95WIVEE528 pKa = 4.24SLLINEE534 pKa = 4.68IEE536 pKa = 4.25SAIYY540 pKa = 10.17RR541 pKa = 11.84SMPMEE546 pKa = 4.98LEE548 pKa = 4.31LDD550 pKa = 3.65AQSEE554 pKa = 4.76YY555 pKa = 10.21TSVMEE560 pKa = 4.0RR561 pKa = 11.84RR562 pKa = 11.84RR563 pKa = 11.84RR564 pKa = 11.84MMLGEE569 pKa = 4.24SVACVDD575 pKa = 3.2WADD578 pKa = 3.9FNITHH583 pKa = 6.27TLKK586 pKa = 11.12DD587 pKa = 3.26MSEE590 pKa = 4.2YY591 pKa = 10.64FLALGEE597 pKa = 4.13AAQRR601 pKa = 11.84ACVEE605 pKa = 4.39NNFVTYY611 pKa = 10.73GGVNYY616 pKa = 10.58GKK618 pKa = 11.15GEE620 pKa = 3.9FLSLCANWCSKK631 pKa = 10.93ALYY634 pKa = 10.62NMWLKK639 pKa = 11.13NGADD643 pKa = 3.51KK644 pKa = 11.3DD645 pKa = 3.89SDD647 pKa = 4.2YY648 pKa = 10.95EE649 pKa = 3.95AHH651 pKa = 7.41ARR653 pKa = 11.84GLWSGWRR660 pKa = 11.84TTSFINCSFNVAYY673 pKa = 10.49CEE675 pKa = 4.61SNFDD679 pKa = 3.91SMHH682 pKa = 6.86HH683 pKa = 6.39IFNSDD688 pKa = 2.96MPLYY692 pKa = 9.86RR693 pKa = 11.84VHH695 pKa = 8.05AGDD698 pKa = 5.47DD699 pKa = 3.67FFGTYY704 pKa = 10.25AEE706 pKa = 5.31EE707 pKa = 3.98IDD709 pKa = 3.3AMRR712 pKa = 11.84FVSSMPLAKK721 pKa = 10.35HH722 pKa = 5.41EE723 pKa = 4.43VNAQKK728 pKa = 10.63QLVGGEE734 pKa = 3.76MGEE737 pKa = 4.3FLRR740 pKa = 11.84HH741 pKa = 5.39EE742 pKa = 4.43YY743 pKa = 10.6FRR745 pKa = 11.84DD746 pKa = 3.47GTVRR750 pKa = 11.84GSVMRR755 pKa = 11.84SIGSFVGSDD764 pKa = 3.29LQAPQLYY771 pKa = 10.37SGVAQCQGANEE782 pKa = 4.96AINLLIRR789 pKa = 11.84RR790 pKa = 11.84GFDD793 pKa = 2.95LRR795 pKa = 11.84AAEE798 pKa = 4.43SIRR801 pKa = 11.84YY802 pKa = 7.67TIVQYY807 pKa = 10.19WGTVKK812 pKa = 10.63EE813 pKa = 4.51PGGSSATPSVKK824 pKa = 10.3LLALSAHH831 pKa = 6.35SGGLGCPRR839 pKa = 11.84FGEE842 pKa = 4.26LATVTRR848 pKa = 11.84EE849 pKa = 3.88RR850 pKa = 11.84LHH852 pKa = 6.83GIPVMRR858 pKa = 11.84DD859 pKa = 3.33VLSKK863 pKa = 10.77LAAPRR868 pKa = 11.84NMEE871 pKa = 4.17RR872 pKa = 11.84LLHH875 pKa = 6.62AFQAKK880 pKa = 8.79IRR882 pKa = 11.84RR883 pKa = 11.84FGYY886 pKa = 9.79IEE888 pKa = 4.06EE889 pKa = 4.87GFTALRR895 pKa = 11.84NAAYY899 pKa = 9.97NATYY903 pKa = 10.52GSDD906 pKa = 3.52LPRR909 pKa = 11.84EE910 pKa = 4.29IKK912 pKa = 10.84LEE914 pKa = 4.05ADD916 pKa = 3.43GEE918 pKa = 4.22LRR920 pKa = 11.84RR921 pKa = 11.84AQVAWINEE929 pKa = 4.29CNLKK933 pKa = 10.0VPEE936 pKa = 4.51MTVAWYY942 pKa = 8.88PSVPSKK948 pKa = 10.69VLEE951 pKa = 3.94YY952 pKa = 10.9LEE954 pKa = 5.17RR955 pKa = 11.84LIACAEE961 pKa = 4.08KK962 pKa = 10.68GLEE965 pKa = 4.19PEE967 pKa = 4.51YY968 pKa = 10.62PMHH971 pKa = 7.34DD972 pKa = 4.01LNGMAVAARR981 pKa = 11.84ATALGAFSALDD992 pKa = 3.23SGLNFLGDD1000 pKa = 3.34EE1001 pKa = 4.52AGRR1004 pKa = 11.84VAGFDD1009 pKa = 3.24AVVAMSRR1016 pKa = 11.84GRR1018 pKa = 11.84AGTILNRR1025 pKa = 11.84MRR1027 pKa = 11.84LYY1029 pKa = 10.75LPEE1032 pKa = 4.8IAVRR1036 pKa = 11.84ALVEE1040 pKa = 4.2HH1041 pKa = 7.04RR1042 pKa = 11.84WNVPTSTGGVVPAQLRR1058 pKa = 11.84GIVFSGLNYY1067 pKa = 10.49ALWYY1071 pKa = 9.23AKK1073 pKa = 10.23NWMPVNAGSLDD1084 pKa = 3.55WLTSYY1089 pKa = 11.14CDD1091 pKa = 3.3AVNIRR1096 pKa = 11.84IANLYY1101 pKa = 10.01KK1102 pKa = 10.64SSLSKK1107 pKa = 9.75MMKK1110 pKa = 8.62MM1111 pKa = 4.54
Molecular weight: 123.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4004 |
227 |
1426 |
1001.0 |
108.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.464 ± 0.555 | 1.374 ± 0.259 |
5.794 ± 0.323 | 4.82 ± 0.51 |
3.571 ± 0.466 | 8.966 ± 0.458 |
1.998 ± 0.306 | 3.796 ± 0.218 |
2.473 ± 0.529 | 9.565 ± 0.777 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.772 ± 0.383 | 4.071 ± 0.669 |
4.121 ± 0.408 | 2.922 ± 0.185 |
7.243 ± 0.165 | 6.069 ± 0.433 |
6.094 ± 0.752 | 8.342 ± 0.786 |
1.499 ± 0.194 | 3.047 ± 0.344 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
