
Leuconostocaceae bacterium R-53105
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; unclassified Lactobacillaceae
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
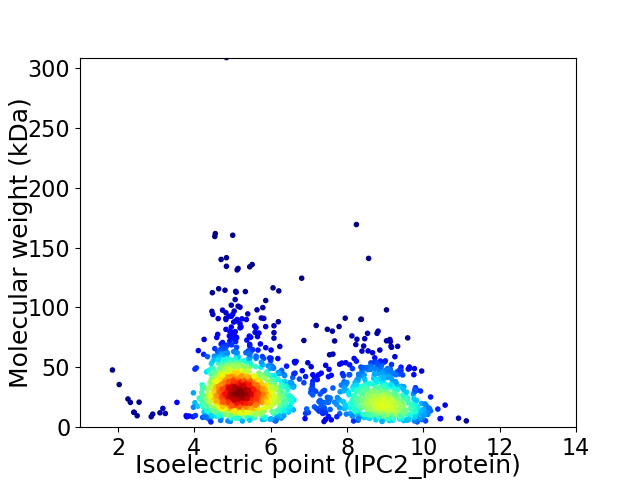
Virtual 2D-PAGE plot for 1574 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G6H475|A0A1G6H475_9LACO ATP-dependent RNA helicase DeaD OS=Leuconostocaceae bacterium R-53105 OX=1855370 GN=SAMN05216341_10341 PE=3 SV=1
MM1 pKa = 7.93EE2 pKa = 6.41DD3 pKa = 3.39NFTTFAKK10 pKa = 10.51SYY12 pKa = 11.46DD13 pKa = 3.73EE14 pKa = 5.98LFDD17 pKa = 4.23NDD19 pKa = 4.94MYY21 pKa = 10.93QAWAQYY27 pKa = 9.97VKK29 pKa = 10.92ALTKK33 pKa = 10.12PSPLLDD39 pKa = 3.91LGGGAGRR46 pKa = 11.84LAVLLEE52 pKa = 3.95QAGYY56 pKa = 10.17QVDD59 pKa = 4.83LLDD62 pKa = 5.04LSPNMLQLAQAHH74 pKa = 5.69AQQAGVDD81 pKa = 3.8LRR83 pKa = 11.84LIEE86 pKa = 4.49GDD88 pKa = 3.39MRR90 pKa = 11.84DD91 pKa = 3.63FSDD94 pKa = 4.1WPTQYY99 pKa = 10.94PIITSFADD107 pKa = 3.29SLNYY111 pKa = 10.32LPNLDD116 pKa = 4.46DD117 pKa = 6.45LEE119 pKa = 5.44DD120 pKa = 3.93AFQQVFDD127 pKa = 4.36HH128 pKa = 6.57LMPSGLFLFDD138 pKa = 4.5VITPYY143 pKa = 10.34QVNVGYY149 pKa = 10.83DD150 pKa = 3.32NYY152 pKa = 11.07CYY154 pKa = 11.1NNDD157 pKa = 4.44DD158 pKa = 3.65NPEE161 pKa = 4.33NIFMWTSFPGEE172 pKa = 4.1NEE174 pKa = 3.65NSVDD178 pKa = 3.43HH179 pKa = 6.86DD180 pKa = 4.46LKK182 pKa = 10.99FFTYY186 pKa = 10.6NEE188 pKa = 3.89EE189 pKa = 3.78LDD191 pKa = 3.73AFNLLRR197 pKa = 11.84EE198 pKa = 4.27VHH200 pKa = 6.6HH201 pKa = 5.93EE202 pKa = 3.83QSYY205 pKa = 9.06EE206 pKa = 3.78QAVYY210 pKa = 10.88CEE212 pKa = 4.55LLQKK216 pKa = 10.86VGFNQIEE223 pKa = 3.89ISADD227 pKa = 3.29FGQTVPNSQTTRR239 pKa = 11.84WFFKK243 pKa = 10.51ASKK246 pKa = 10.51GDD248 pKa = 3.56TKK250 pKa = 11.37
MM1 pKa = 7.93EE2 pKa = 6.41DD3 pKa = 3.39NFTTFAKK10 pKa = 10.51SYY12 pKa = 11.46DD13 pKa = 3.73EE14 pKa = 5.98LFDD17 pKa = 4.23NDD19 pKa = 4.94MYY21 pKa = 10.93QAWAQYY27 pKa = 9.97VKK29 pKa = 10.92ALTKK33 pKa = 10.12PSPLLDD39 pKa = 3.91LGGGAGRR46 pKa = 11.84LAVLLEE52 pKa = 3.95QAGYY56 pKa = 10.17QVDD59 pKa = 4.83LLDD62 pKa = 5.04LSPNMLQLAQAHH74 pKa = 5.69AQQAGVDD81 pKa = 3.8LRR83 pKa = 11.84LIEE86 pKa = 4.49GDD88 pKa = 3.39MRR90 pKa = 11.84DD91 pKa = 3.63FSDD94 pKa = 4.1WPTQYY99 pKa = 10.94PIITSFADD107 pKa = 3.29SLNYY111 pKa = 10.32LPNLDD116 pKa = 4.46DD117 pKa = 6.45LEE119 pKa = 5.44DD120 pKa = 3.93AFQQVFDD127 pKa = 4.36HH128 pKa = 6.57LMPSGLFLFDD138 pKa = 4.5VITPYY143 pKa = 10.34QVNVGYY149 pKa = 10.83DD150 pKa = 3.32NYY152 pKa = 11.07CYY154 pKa = 11.1NNDD157 pKa = 4.44DD158 pKa = 3.65NPEE161 pKa = 4.33NIFMWTSFPGEE172 pKa = 4.1NEE174 pKa = 3.65NSVDD178 pKa = 3.43HH179 pKa = 6.86DD180 pKa = 4.46LKK182 pKa = 10.99FFTYY186 pKa = 10.6NEE188 pKa = 3.89EE189 pKa = 3.78LDD191 pKa = 3.73AFNLLRR197 pKa = 11.84EE198 pKa = 4.27VHH200 pKa = 6.6HH201 pKa = 5.93EE202 pKa = 3.83QSYY205 pKa = 9.06EE206 pKa = 3.78QAVYY210 pKa = 10.88CEE212 pKa = 4.55LLQKK216 pKa = 10.86VGFNQIEE223 pKa = 3.89ISADD227 pKa = 3.29FGQTVPNSQTTRR239 pKa = 11.84WFFKK243 pKa = 10.51ASKK246 pKa = 10.51GDD248 pKa = 3.56TKK250 pKa = 11.37
Molecular weight: 28.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G6IRS7|A0A1G6IRS7_9LACO Acetyl esterase/lipase OS=Leuconostocaceae bacterium R-53105 OX=1855370 GN=SAMN05216341_11215 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.19KK9 pKa = 7.37RR10 pKa = 11.84HH11 pKa = 5.5RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 5.84GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.23VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.19KK9 pKa = 7.37RR10 pKa = 11.84HH11 pKa = 5.5RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 5.84GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 8.23VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
476006 |
39 |
2928 |
302.4 |
33.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.861 ± 0.09 | 0.265 ± 0.011 |
5.886 ± 0.06 | 5.228 ± 0.07 |
4.131 ± 0.056 | 6.651 ± 0.061 |
2.008 ± 0.026 | 6.993 ± 0.074 |
5.608 ± 0.065 | 10.13 ± 0.099 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.515 ± 0.035 | 4.802 ± 0.05 |
3.653 ± 0.038 | 5.749 ± 0.079 |
3.903 ± 0.052 | 6.266 ± 0.133 |
5.842 ± 0.062 | 7.119 ± 0.058 |
1.071 ± 0.026 | 3.318 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
