
Bacillus sp. AFS006103
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Bacillus; unclassified Bacillus (in: Bacteria)
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
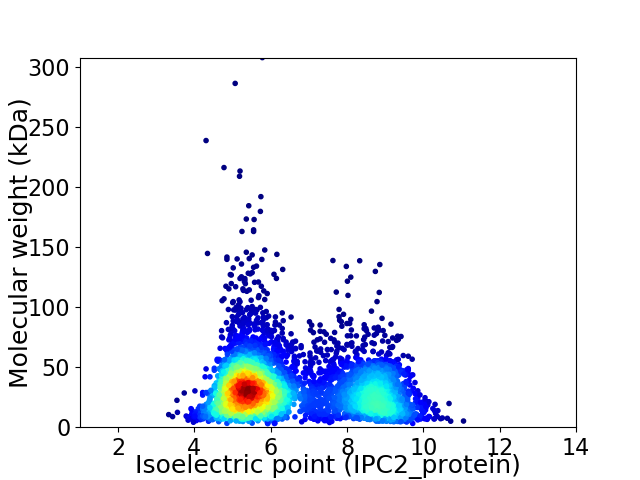
Virtual 2D-PAGE plot for 4840 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A8IEV9|A0A2A8IEV9_9BACI Uncharacterized protein OS=Bacillus sp. AFS006103 OX=2033483 GN=CN481_15035 PE=4 SV=1
MM1 pKa = 7.5SLEE4 pKa = 3.92WFDD7 pKa = 4.01RR8 pKa = 11.84VCGEE12 pKa = 4.07LQEE15 pKa = 4.49HH16 pKa = 6.69LEE18 pKa = 4.58SICEE22 pKa = 4.09EE23 pKa = 4.21YY24 pKa = 11.03DD25 pKa = 3.13QVGQMTVEE33 pKa = 4.33RR34 pKa = 11.84AAKK37 pKa = 9.24HH38 pKa = 5.09PRR40 pKa = 11.84IEE42 pKa = 4.68FYY44 pKa = 11.41VDD46 pKa = 2.79TADD49 pKa = 3.6VDD51 pKa = 4.09TEE53 pKa = 4.13EE54 pKa = 5.17LEE56 pKa = 4.19RR57 pKa = 11.84DD58 pKa = 3.96YY59 pKa = 11.3FCSLFFDD66 pKa = 4.14PQNEE70 pKa = 3.98EE71 pKa = 4.38FYY73 pKa = 10.64IDD75 pKa = 3.79TFDD78 pKa = 3.5VDD80 pKa = 3.7SGHH83 pKa = 5.15TAKK86 pKa = 10.89VILSDD91 pKa = 3.3IEE93 pKa = 5.03DD94 pKa = 4.03IIEE97 pKa = 4.01EE98 pKa = 4.26VHH100 pKa = 7.07ASLHH104 pKa = 7.05DD105 pKa = 3.91YY106 pKa = 10.93MNGDD110 pKa = 3.64DD111 pKa = 5.48HH112 pKa = 6.4YY113 pKa = 9.46TDD115 pKa = 4.67DD116 pKa = 4.74GVYY119 pKa = 10.52LQTTEE124 pKa = 4.76VSDD127 pKa = 4.83EE128 pKa = 4.12DD129 pKa = 5.49DD130 pKa = 3.44EE131 pKa = 5.59DD132 pKa = 5.2NEE134 pKa = 4.33VEE136 pKa = 4.37TVYY139 pKa = 11.13VEE141 pKa = 4.55DD142 pKa = 6.0DD143 pKa = 3.5EE144 pKa = 6.45DD145 pKa = 3.85EE146 pKa = 4.62EE147 pKa = 5.04YY148 pKa = 11.52YY149 pKa = 10.68EE150 pKa = 4.27AAQVNSDD157 pKa = 4.91DD158 pKa = 3.62IFEE161 pKa = 5.14EE162 pKa = 4.61IDD164 pKa = 3.7VDD166 pKa = 3.35WSTPEE171 pKa = 3.51VTAFKK176 pKa = 10.83HH177 pKa = 5.1EE178 pKa = 4.6DD179 pKa = 3.76EE180 pKa = 5.29VEE182 pKa = 3.93VTYY185 pKa = 10.99QFGLVQATGDD195 pKa = 3.65GVLKK199 pKa = 10.42RR200 pKa = 11.84INRR203 pKa = 11.84IWTTDD208 pKa = 3.36DD209 pKa = 3.95EE210 pKa = 4.93LIKK213 pKa = 11.03DD214 pKa = 3.59EE215 pKa = 4.46SHH217 pKa = 7.38FIFSKK222 pKa = 11.09EE223 pKa = 3.67EE224 pKa = 3.68ATTIIAMIASHH235 pKa = 6.76MDD237 pKa = 3.18QLSEE241 pKa = 4.65FEE243 pKa = 4.27FF244 pKa = 5.59
MM1 pKa = 7.5SLEE4 pKa = 3.92WFDD7 pKa = 4.01RR8 pKa = 11.84VCGEE12 pKa = 4.07LQEE15 pKa = 4.49HH16 pKa = 6.69LEE18 pKa = 4.58SICEE22 pKa = 4.09EE23 pKa = 4.21YY24 pKa = 11.03DD25 pKa = 3.13QVGQMTVEE33 pKa = 4.33RR34 pKa = 11.84AAKK37 pKa = 9.24HH38 pKa = 5.09PRR40 pKa = 11.84IEE42 pKa = 4.68FYY44 pKa = 11.41VDD46 pKa = 2.79TADD49 pKa = 3.6VDD51 pKa = 4.09TEE53 pKa = 4.13EE54 pKa = 5.17LEE56 pKa = 4.19RR57 pKa = 11.84DD58 pKa = 3.96YY59 pKa = 11.3FCSLFFDD66 pKa = 4.14PQNEE70 pKa = 3.98EE71 pKa = 4.38FYY73 pKa = 10.64IDD75 pKa = 3.79TFDD78 pKa = 3.5VDD80 pKa = 3.7SGHH83 pKa = 5.15TAKK86 pKa = 10.89VILSDD91 pKa = 3.3IEE93 pKa = 5.03DD94 pKa = 4.03IIEE97 pKa = 4.01EE98 pKa = 4.26VHH100 pKa = 7.07ASLHH104 pKa = 7.05DD105 pKa = 3.91YY106 pKa = 10.93MNGDD110 pKa = 3.64DD111 pKa = 5.48HH112 pKa = 6.4YY113 pKa = 9.46TDD115 pKa = 4.67DD116 pKa = 4.74GVYY119 pKa = 10.52LQTTEE124 pKa = 4.76VSDD127 pKa = 4.83EE128 pKa = 4.12DD129 pKa = 5.49DD130 pKa = 3.44EE131 pKa = 5.59DD132 pKa = 5.2NEE134 pKa = 4.33VEE136 pKa = 4.37TVYY139 pKa = 11.13VEE141 pKa = 4.55DD142 pKa = 6.0DD143 pKa = 3.5EE144 pKa = 6.45DD145 pKa = 3.85EE146 pKa = 4.62EE147 pKa = 5.04YY148 pKa = 11.52YY149 pKa = 10.68EE150 pKa = 4.27AAQVNSDD157 pKa = 4.91DD158 pKa = 3.62IFEE161 pKa = 5.14EE162 pKa = 4.61IDD164 pKa = 3.7VDD166 pKa = 3.35WSTPEE171 pKa = 3.51VTAFKK176 pKa = 10.83HH177 pKa = 5.1EE178 pKa = 4.6DD179 pKa = 3.76EE180 pKa = 5.29VEE182 pKa = 3.93VTYY185 pKa = 10.99QFGLVQATGDD195 pKa = 3.65GVLKK199 pKa = 10.42RR200 pKa = 11.84INRR203 pKa = 11.84IWTTDD208 pKa = 3.36DD209 pKa = 3.95EE210 pKa = 4.93LIKK213 pKa = 11.03DD214 pKa = 3.59EE215 pKa = 4.46SHH217 pKa = 7.38FIFSKK222 pKa = 11.09EE223 pKa = 3.67EE224 pKa = 3.68ATTIIAMIASHH235 pKa = 6.76MDD237 pKa = 3.18QLSEE241 pKa = 4.65FEE243 pKa = 4.27FF244 pKa = 5.59
Molecular weight: 28.48 kDa
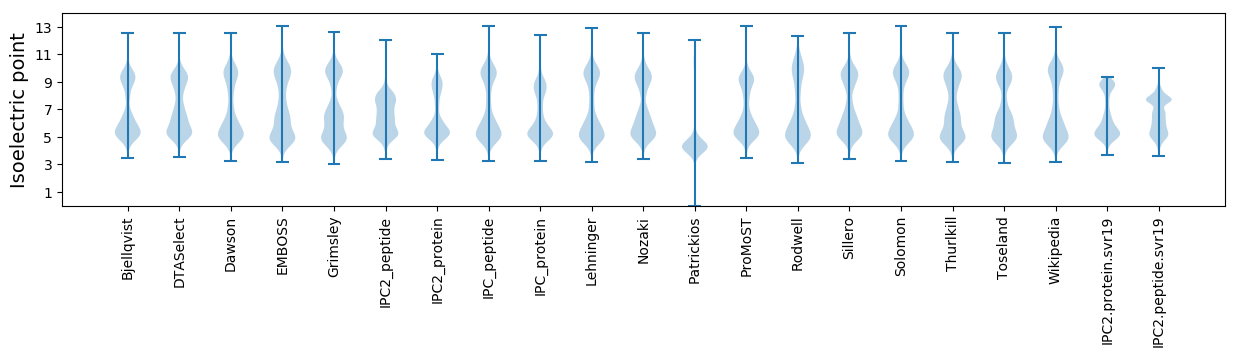
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A8IMB0|A0A2A8IMB0_9BACI Phosphate-specific transport system accessory protein PhoU OS=Bacillus sp. AFS006103 OX=2033483 GN=phoU PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.38RR3 pKa = 11.84TYY5 pKa = 10.17QPNSRR10 pKa = 11.84KK11 pKa = 9.39HH12 pKa = 5.91SKK14 pKa = 9.08VHH16 pKa = 5.89GFRR19 pKa = 11.84SRR21 pKa = 11.84MSSANGRR28 pKa = 11.84KK29 pKa = 8.77VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.43GRR39 pKa = 11.84KK40 pKa = 8.78VLSAA44 pKa = 4.05
MM1 pKa = 7.35KK2 pKa = 9.38RR3 pKa = 11.84TYY5 pKa = 10.17QPNSRR10 pKa = 11.84KK11 pKa = 9.39HH12 pKa = 5.91SKK14 pKa = 9.08VHH16 pKa = 5.89GFRR19 pKa = 11.84SRR21 pKa = 11.84MSSANGRR28 pKa = 11.84KK29 pKa = 8.77VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.43GRR39 pKa = 11.84KK40 pKa = 8.78VLSAA44 pKa = 4.05
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1451607 |
27 |
2706 |
299.9 |
33.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.8 ± 0.041 | 0.781 ± 0.012 |
4.89 ± 0.026 | 7.183 ± 0.039 |
4.683 ± 0.027 | 6.94 ± 0.037 |
2.018 ± 0.016 | 8.073 ± 0.038 |
7.227 ± 0.038 | 9.876 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.714 ± 0.018 | 4.686 ± 0.026 |
3.638 ± 0.022 | 3.65 ± 0.022 |
3.923 ± 0.025 | 6.05 ± 0.025 |
5.414 ± 0.031 | 6.877 ± 0.028 |
1.044 ± 0.013 | 3.533 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
