
Domestic cat hepadnavirus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Blubervirales; Hepadnaviridae; Orthohepadnavirus; Domestic cat hepatitis B virus
Average proteome isoelectric point is 8.71
Get precalculated fractions of proteins
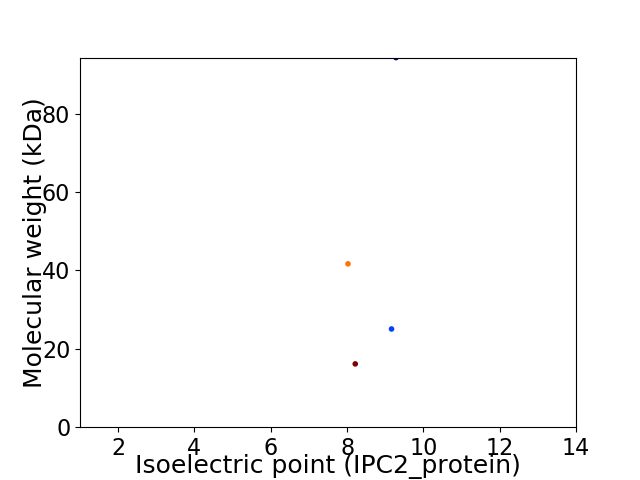
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U8SZQ3|A0A2U8SZQ3_9HEPA External core antigen OS=Domestic cat hepadnavirus OX=2107574 GN=C PE=3 SV=1
MM1 pKa = 7.64GANQSIPNPLGFFPRR16 pKa = 11.84HH17 pKa = 5.71NLDD20 pKa = 3.23LTDD23 pKa = 4.0PAGTLGDD30 pKa = 4.14FDD32 pKa = 5.88HH33 pKa = 7.12NPLKK37 pKa = 10.88DD38 pKa = 3.41PWPQSSHH45 pKa = 5.01QSRR48 pKa = 11.84GAWGPGFVPPHH59 pKa = 6.31GGLLGSLSFATQGVVSAQPITNPPPRR85 pKa = 11.84KK86 pKa = 9.28KK87 pKa = 10.85GRR89 pKa = 11.84GPTPLTPPVRR99 pKa = 11.84VTHH102 pKa = 6.24PQMSPSSWRR111 pKa = 11.84QKK113 pKa = 10.61YY114 pKa = 10.42GYY116 pKa = 10.42LLLPKK121 pKa = 8.28TTAAPGPTSPPQTTVAPTASSTSSPPLTIGAPAARR156 pKa = 11.84MGNITSGPFGFLLGLQVGSFLWTKK180 pKa = 9.9IQTIGQSADD189 pKa = 3.32SWWTSLSFPGAIPGCIGQDD208 pKa = 3.2SQYY211 pKa = 9.22QTSKK215 pKa = 10.67HH216 pKa = 6.39SPTSCPPTCTGFPWMCLRR234 pKa = 11.84RR235 pKa = 11.84FIIYY239 pKa = 10.6LLLLALLLIFSLVLLDD255 pKa = 3.78WKK257 pKa = 11.0GLLPVCPFPPGDD269 pKa = 3.82SQTGNIRR276 pKa = 11.84CKK278 pKa = 10.64ACIGSAPEE286 pKa = 4.33TPWPPLCCCTVPSDD300 pKa = 4.59GNCTCWPIPSSWALGSYY317 pKa = 9.68LWEE320 pKa = 4.06LALARR325 pKa = 11.84FSWLSSLVVWLQWLGGISPIVWCLLIWMTWFWGLRR360 pKa = 11.84VWNILSQFMPLLFLLFYY377 pKa = 10.7HH378 pKa = 7.07LVYY381 pKa = 10.34TT382 pKa = 4.99
MM1 pKa = 7.64GANQSIPNPLGFFPRR16 pKa = 11.84HH17 pKa = 5.71NLDD20 pKa = 3.23LTDD23 pKa = 4.0PAGTLGDD30 pKa = 4.14FDD32 pKa = 5.88HH33 pKa = 7.12NPLKK37 pKa = 10.88DD38 pKa = 3.41PWPQSSHH45 pKa = 5.01QSRR48 pKa = 11.84GAWGPGFVPPHH59 pKa = 6.31GGLLGSLSFATQGVVSAQPITNPPPRR85 pKa = 11.84KK86 pKa = 9.28KK87 pKa = 10.85GRR89 pKa = 11.84GPTPLTPPVRR99 pKa = 11.84VTHH102 pKa = 6.24PQMSPSSWRR111 pKa = 11.84QKK113 pKa = 10.61YY114 pKa = 10.42GYY116 pKa = 10.42LLLPKK121 pKa = 8.28TTAAPGPTSPPQTTVAPTASSTSSPPLTIGAPAARR156 pKa = 11.84MGNITSGPFGFLLGLQVGSFLWTKK180 pKa = 9.9IQTIGQSADD189 pKa = 3.32SWWTSLSFPGAIPGCIGQDD208 pKa = 3.2SQYY211 pKa = 9.22QTSKK215 pKa = 10.67HH216 pKa = 6.39SPTSCPPTCTGFPWMCLRR234 pKa = 11.84RR235 pKa = 11.84FIIYY239 pKa = 10.6LLLLALLLIFSLVLLDD255 pKa = 3.78WKK257 pKa = 11.0GLLPVCPFPPGDD269 pKa = 3.82SQTGNIRR276 pKa = 11.84CKK278 pKa = 10.64ACIGSAPEE286 pKa = 4.33TPWPPLCCCTVPSDD300 pKa = 4.59GNCTCWPIPSSWALGSYY317 pKa = 9.68LWEE320 pKa = 4.06LALARR325 pKa = 11.84FSWLSSLVVWLQWLGGISPIVWCLLIWMTWFWGLRR360 pKa = 11.84VWNILSQFMPLLFLLFYY377 pKa = 10.7HH378 pKa = 7.07LVYY381 pKa = 10.34TT382 pKa = 4.99
Molecular weight: 41.66 kDa
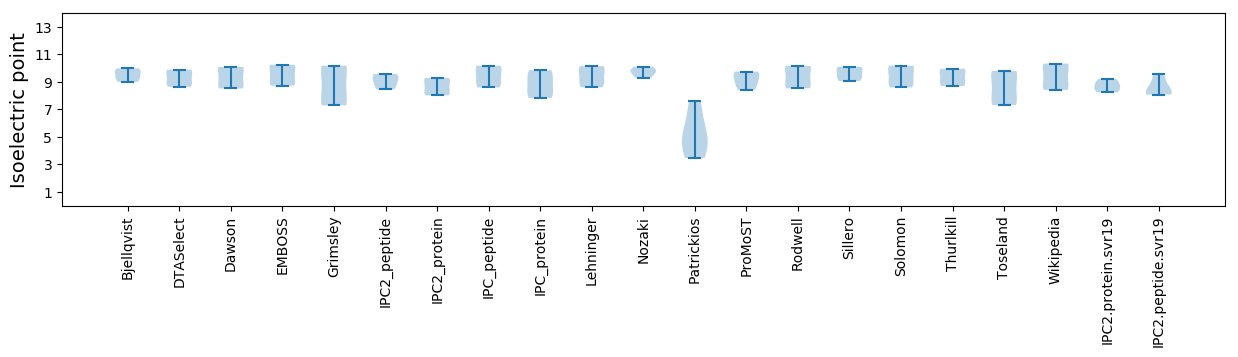
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U8T025|A0A2U8T025_9HEPA Protein X OS=Domestic cat hepadnavirus OX=2107574 GN=X PE=3 SV=1
MM1 pKa = 7.47HH2 pKa = 7.68LFHH5 pKa = 7.08LCVLFCSIPTVQASKK20 pKa = 11.21LCLRR24 pKa = 11.84WLWGMDD30 pKa = 2.7IDD32 pKa = 4.44PYY34 pKa = 11.38KK35 pKa = 10.94EE36 pKa = 4.02FGTTSQLISFLPSDD50 pKa = 4.81FFPALNDD57 pKa = 3.91LVDD60 pKa = 4.86TIQALYY66 pKa = 10.59EE67 pKa = 4.01EE68 pKa = 5.04EE69 pKa = 4.11LTGRR73 pKa = 11.84EE74 pKa = 4.29HH75 pKa = 7.77CSPHH79 pKa = 5.03HH80 pKa = 5.16TALRR84 pKa = 11.84VLLNCWEE91 pKa = 4.24EE92 pKa = 4.25SARR95 pKa = 11.84MATWVRR101 pKa = 11.84ANVEE105 pKa = 4.1GAPLQDD111 pKa = 4.8AIVAYY116 pKa = 10.69VNSTVSLKK124 pKa = 10.44LRR126 pKa = 11.84QQMWFHH132 pKa = 6.56LSCLTFGQHH141 pKa = 5.14TVLEE145 pKa = 4.45FLVSFGTWIRR155 pKa = 11.84TPAPYY160 pKa = 9.94RR161 pKa = 11.84PPNAPILSTLPEE173 pKa = 4.04HH174 pKa = 5.47TVIRR178 pKa = 11.84ARR180 pKa = 11.84GAARR184 pKa = 11.84RR185 pKa = 11.84PARR188 pKa = 11.84SPRR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84TPSPRR198 pKa = 11.84RR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84SQSPRR206 pKa = 11.84RR207 pKa = 11.84RR208 pKa = 11.84RR209 pKa = 11.84SQSPTQSNCC218 pKa = 2.89
MM1 pKa = 7.47HH2 pKa = 7.68LFHH5 pKa = 7.08LCVLFCSIPTVQASKK20 pKa = 11.21LCLRR24 pKa = 11.84WLWGMDD30 pKa = 2.7IDD32 pKa = 4.44PYY34 pKa = 11.38KK35 pKa = 10.94EE36 pKa = 4.02FGTTSQLISFLPSDD50 pKa = 4.81FFPALNDD57 pKa = 3.91LVDD60 pKa = 4.86TIQALYY66 pKa = 10.59EE67 pKa = 4.01EE68 pKa = 5.04EE69 pKa = 4.11LTGRR73 pKa = 11.84EE74 pKa = 4.29HH75 pKa = 7.77CSPHH79 pKa = 5.03HH80 pKa = 5.16TALRR84 pKa = 11.84VLLNCWEE91 pKa = 4.24EE92 pKa = 4.25SARR95 pKa = 11.84MATWVRR101 pKa = 11.84ANVEE105 pKa = 4.1GAPLQDD111 pKa = 4.8AIVAYY116 pKa = 10.69VNSTVSLKK124 pKa = 10.44LRR126 pKa = 11.84QQMWFHH132 pKa = 6.56LSCLTFGQHH141 pKa = 5.14TVLEE145 pKa = 4.45FLVSFGTWIRR155 pKa = 11.84TPAPYY160 pKa = 9.94RR161 pKa = 11.84PPNAPILSTLPEE173 pKa = 4.04HH174 pKa = 5.47TVIRR178 pKa = 11.84ARR180 pKa = 11.84GAARR184 pKa = 11.84RR185 pKa = 11.84PARR188 pKa = 11.84SPRR191 pKa = 11.84RR192 pKa = 11.84RR193 pKa = 11.84TPSPRR198 pKa = 11.84RR199 pKa = 11.84RR200 pKa = 11.84RR201 pKa = 11.84SQSPRR206 pKa = 11.84RR207 pKa = 11.84RR208 pKa = 11.84RR209 pKa = 11.84SQSPTQSNCC218 pKa = 2.89
Molecular weight: 25.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1583 |
145 |
838 |
395.8 |
44.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.822 ± 0.456 | 2.78 ± 0.551 |
3.032 ± 0.165 | 2.716 ± 0.665 |
4.991 ± 0.573 | 6.886 ± 0.791 |
3.538 ± 0.75 | 3.095 ± 0.532 |
2.59 ± 0.57 | 12.697 ± 0.538 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.832 ± 0.078 | 2.527 ± 0.4 |
8.97 ± 1.11 | 3.79 ± 0.33 |
7.202 ± 1.23 | 9.16 ± 0.419 |
6.191 ± 1.025 | 5.433 ± 0.525 |
3.095 ± 0.697 | 2.653 ± 0.672 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
