
Olsenella sp. An188
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Coriobacteriia; Coriobacteriales; Atopobiaceae; Olsenella; unclassified Olsenella
Average proteome isoelectric point is 5.64
Get precalculated fractions of proteins
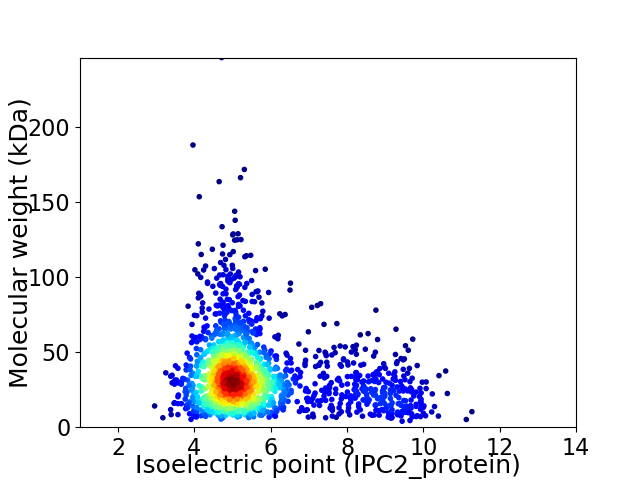
Virtual 2D-PAGE plot for 1814 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
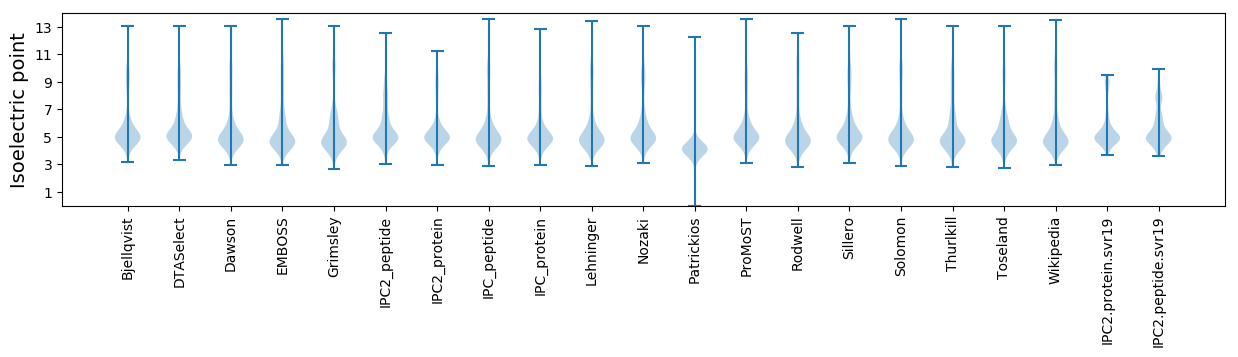
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4K9Q7|A0A1Y4K9Q7_9ACTN Dihydrodipicolinate synthase family protein OS=Olsenella sp. An188 OX=1965579 GN=B5F23_01925 PE=3 SV=1
MM1 pKa = 7.38SNISRR6 pKa = 11.84RR7 pKa = 11.84TFLAGSSASLLAVFLGGCTGSNEE30 pKa = 3.82PAAEE34 pKa = 4.36GEE36 pKa = 4.43GTEE39 pKa = 4.25AAAGGHH45 pKa = 6.24IYY47 pKa = 10.97VLTASPDD54 pKa = 3.67HH55 pKa = 6.45GWTGQVGVFAQEE67 pKa = 4.24KK68 pKa = 8.42VDD70 pKa = 3.8EE71 pKa = 4.37VNEE74 pKa = 4.3AGVYY78 pKa = 7.97TAEE81 pKa = 4.31LQTAGAAADD90 pKa = 3.83QIAQVEE96 pKa = 4.67DD97 pKa = 3.61ILAGDD102 pKa = 4.33LSGVAGIVIQPLDD115 pKa = 3.95DD116 pKa = 3.77TVQSAIQQIVDD127 pKa = 3.13AGVAYY132 pKa = 10.52VAFDD136 pKa = 4.56RR137 pKa = 11.84IIEE140 pKa = 4.23AVQDD144 pKa = 3.67SAVANVKK151 pKa = 10.13GDD153 pKa = 3.47NSGIGAGSAAYY164 pKa = 9.38FVEE167 pKa = 4.77NGMKK171 pKa = 10.29PGDD174 pKa = 3.48AVYY177 pKa = 9.8IYY179 pKa = 10.58QGDD182 pKa = 3.95TSSVTTLRR190 pKa = 11.84DD191 pKa = 3.34NGFTDD196 pKa = 3.72YY197 pKa = 11.7LLGNLEE203 pKa = 4.05FDD205 pKa = 3.81GATVPEE211 pKa = 4.37DD212 pKa = 3.64AKK214 pKa = 9.42WTQEE218 pKa = 4.14QIDD221 pKa = 3.97SAITKK226 pKa = 9.71SGAMNWSRR234 pKa = 11.84SDD236 pKa = 3.05TKK238 pKa = 11.18AAFEE242 pKa = 4.35ALLGDD247 pKa = 4.08EE248 pKa = 5.03ANASIKK254 pKa = 8.48WWYY257 pKa = 10.82AEE259 pKa = 4.09DD260 pKa = 5.16DD261 pKa = 4.0EE262 pKa = 5.02LAMGILEE269 pKa = 4.42ALNGGGISDD278 pKa = 3.84SAKK281 pKa = 8.93EE282 pKa = 3.77AFYY285 pKa = 9.38ATKK288 pKa = 10.34PFITGCGGLQEE299 pKa = 4.55YY300 pKa = 10.58YY301 pKa = 10.84DD302 pKa = 4.27VISGEE307 pKa = 4.49SYY309 pKa = 11.26TDD311 pKa = 2.86ITANLGGVMSTTYY324 pKa = 10.04SPSMIQTAIQDD335 pKa = 4.12MVDD338 pKa = 3.37HH339 pKa = 7.28LDD341 pKa = 3.86GKK343 pKa = 9.62EE344 pKa = 3.92VPQDD348 pKa = 3.41HH349 pKa = 7.11VIACEE354 pKa = 3.73NVTAEE359 pKa = 4.12NVADD363 pKa = 3.85YY364 pKa = 11.46VGFEE368 pKa = 3.98
MM1 pKa = 7.38SNISRR6 pKa = 11.84RR7 pKa = 11.84TFLAGSSASLLAVFLGGCTGSNEE30 pKa = 3.82PAAEE34 pKa = 4.36GEE36 pKa = 4.43GTEE39 pKa = 4.25AAAGGHH45 pKa = 6.24IYY47 pKa = 10.97VLTASPDD54 pKa = 3.67HH55 pKa = 6.45GWTGQVGVFAQEE67 pKa = 4.24KK68 pKa = 8.42VDD70 pKa = 3.8EE71 pKa = 4.37VNEE74 pKa = 4.3AGVYY78 pKa = 7.97TAEE81 pKa = 4.31LQTAGAAADD90 pKa = 3.83QIAQVEE96 pKa = 4.67DD97 pKa = 3.61ILAGDD102 pKa = 4.33LSGVAGIVIQPLDD115 pKa = 3.95DD116 pKa = 3.77TVQSAIQQIVDD127 pKa = 3.13AGVAYY132 pKa = 10.52VAFDD136 pKa = 4.56RR137 pKa = 11.84IIEE140 pKa = 4.23AVQDD144 pKa = 3.67SAVANVKK151 pKa = 10.13GDD153 pKa = 3.47NSGIGAGSAAYY164 pKa = 9.38FVEE167 pKa = 4.77NGMKK171 pKa = 10.29PGDD174 pKa = 3.48AVYY177 pKa = 9.8IYY179 pKa = 10.58QGDD182 pKa = 3.95TSSVTTLRR190 pKa = 11.84DD191 pKa = 3.34NGFTDD196 pKa = 3.72YY197 pKa = 11.7LLGNLEE203 pKa = 4.05FDD205 pKa = 3.81GATVPEE211 pKa = 4.37DD212 pKa = 3.64AKK214 pKa = 9.42WTQEE218 pKa = 4.14QIDD221 pKa = 3.97SAITKK226 pKa = 9.71SGAMNWSRR234 pKa = 11.84SDD236 pKa = 3.05TKK238 pKa = 11.18AAFEE242 pKa = 4.35ALLGDD247 pKa = 4.08EE248 pKa = 5.03ANASIKK254 pKa = 8.48WWYY257 pKa = 10.82AEE259 pKa = 4.09DD260 pKa = 5.16DD261 pKa = 4.0EE262 pKa = 5.02LAMGILEE269 pKa = 4.42ALNGGGISDD278 pKa = 3.84SAKK281 pKa = 8.93EE282 pKa = 3.77AFYY285 pKa = 9.38ATKK288 pKa = 10.34PFITGCGGLQEE299 pKa = 4.55YY300 pKa = 10.58YY301 pKa = 10.84DD302 pKa = 4.27VISGEE307 pKa = 4.49SYY309 pKa = 11.26TDD311 pKa = 2.86ITANLGGVMSTTYY324 pKa = 10.04SPSMIQTAIQDD335 pKa = 4.12MVDD338 pKa = 3.37HH339 pKa = 7.28LDD341 pKa = 3.86GKK343 pKa = 9.62EE344 pKa = 3.92VPQDD348 pKa = 3.41HH349 pKa = 7.11VIACEE354 pKa = 3.73NVTAEE359 pKa = 4.12NVADD363 pKa = 3.85YY364 pKa = 11.46VGFEE368 pKa = 3.98
Molecular weight: 38.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4K5A3|A0A1Y4K5A3_9ACTN Uncharacterized protein OS=Olsenella sp. An188 OX=1965579 GN=B5F23_04275 PE=4 SV=1
MM1 pKa = 7.48TFGTTTPRR9 pKa = 11.84PRR11 pKa = 11.84PPSRR15 pKa = 11.84ARR17 pKa = 11.84ARR19 pKa = 11.84VRR21 pKa = 11.84GRR23 pKa = 11.84TSRR26 pKa = 11.84SPRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84SRR34 pKa = 11.84RR35 pKa = 11.84TAPPPPRR42 pKa = 11.84GCRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84SRR51 pKa = 11.84RR52 pKa = 11.84SRR54 pKa = 11.84CRR56 pKa = 11.84RR57 pKa = 11.84WAPRR61 pKa = 11.84RR62 pKa = 11.84TPAQTSRR69 pKa = 11.84GPRR72 pKa = 11.84RR73 pKa = 11.84PRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84ATAGPRR85 pKa = 11.84SGG87 pKa = 3.92
MM1 pKa = 7.48TFGTTTPRR9 pKa = 11.84PRR11 pKa = 11.84PPSRR15 pKa = 11.84ARR17 pKa = 11.84ARR19 pKa = 11.84VRR21 pKa = 11.84GRR23 pKa = 11.84TSRR26 pKa = 11.84SPRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84SRR34 pKa = 11.84RR35 pKa = 11.84TAPPPPRR42 pKa = 11.84GCRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84SRR51 pKa = 11.84RR52 pKa = 11.84SRR54 pKa = 11.84CRR56 pKa = 11.84RR57 pKa = 11.84WAPRR61 pKa = 11.84RR62 pKa = 11.84TPAQTSRR69 pKa = 11.84GPRR72 pKa = 11.84RR73 pKa = 11.84PRR75 pKa = 11.84RR76 pKa = 11.84RR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84ATAGPRR85 pKa = 11.84SGG87 pKa = 3.92
Molecular weight: 10.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
615698 |
36 |
2282 |
339.4 |
36.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.452 ± 0.084 | 1.517 ± 0.024 |
6.259 ± 0.043 | 7.062 ± 0.049 |
3.286 ± 0.034 | 8.786 ± 0.053 |
1.809 ± 0.023 | 3.942 ± 0.053 |
2.63 ± 0.046 | 9.684 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.394 ± 0.027 | 2.327 ± 0.032 |
4.436 ± 0.035 | 2.467 ± 0.03 |
7.27 ± 0.07 | 5.744 ± 0.041 |
5.157 ± 0.052 | 9.042 ± 0.051 |
1.113 ± 0.019 | 2.623 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
