
Beihai noda-like virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
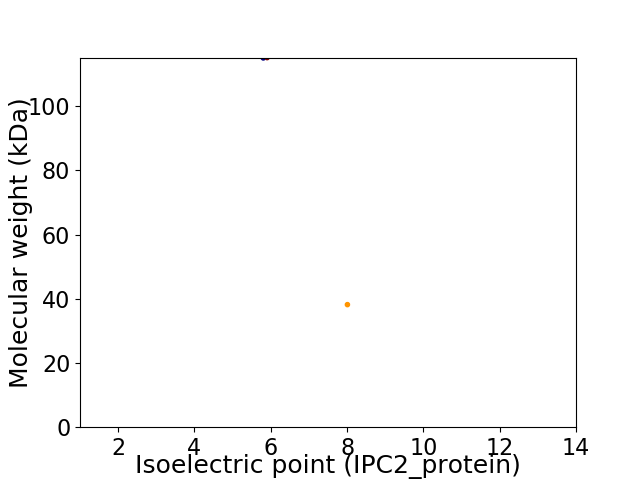
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFY0|A0A1L3KFY0_9VIRU RNA replicase OS=Beihai noda-like virus 5 OX=1922487 PE=3 SV=1
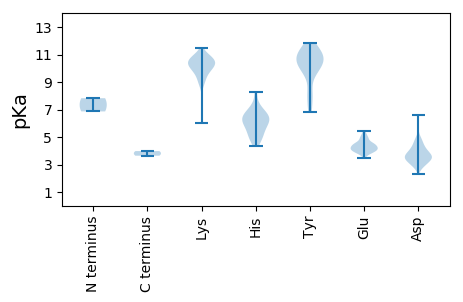
MM1 pKa = 7.82LSRR4 pKa = 11.84LYY6 pKa = 10.55HH7 pKa = 5.19VVEE10 pKa = 4.42EE11 pKa = 4.56EE12 pKa = 4.9VFEE15 pKa = 4.33PWAKK19 pKa = 10.6ALARR23 pKa = 11.84PVLHH27 pKa = 6.24VAHH30 pKa = 6.65PVVTAAYY37 pKa = 9.62DD38 pKa = 3.43VTDD41 pKa = 3.67QVSRR45 pKa = 11.84WLGPGYY51 pKa = 7.9GTCPPLSPAEE61 pKa = 3.93EE62 pKa = 3.89VVAFIRR68 pKa = 11.84SCWPDD73 pKa = 3.62LEE75 pKa = 4.57QIDD78 pKa = 4.15YY79 pKa = 11.17ALLIGCRR86 pKa = 11.84LVLLIAGAVILFNLQRR102 pKa = 11.84KK103 pKa = 6.0VLRR106 pKa = 11.84TISITNPVRR115 pKa = 11.84QYY117 pKa = 11.21VIEE120 pKa = 4.06LSKK123 pKa = 10.63IKK125 pKa = 10.23QYY127 pKa = 11.08PDD129 pKa = 3.42DD130 pKa = 3.83LRR132 pKa = 11.84KK133 pKa = 9.94KK134 pKa = 10.04FRR136 pKa = 11.84NLNVLKK142 pKa = 10.36YY143 pKa = 9.65APSLNGHH150 pKa = 5.11SHH152 pKa = 6.1GVCAAFRR159 pKa = 11.84AAADD163 pKa = 3.55DD164 pKa = 4.33TINTFIQEE172 pKa = 4.04NGFEE176 pKa = 4.34VYY178 pKa = 10.31SVGMSAKK185 pKa = 10.8DD186 pKa = 3.19MDD188 pKa = 4.82LGFEE192 pKa = 5.03GYY194 pKa = 10.49RR195 pKa = 11.84PMVEE199 pKa = 4.26DD200 pKa = 4.08KK201 pKa = 11.51DD202 pKa = 3.57MVMGFKK208 pKa = 10.51CSPVRR213 pKa = 11.84DD214 pKa = 3.49NHH216 pKa = 6.17IIKK219 pKa = 9.63MVDD222 pKa = 2.31VDD224 pKa = 4.24YY225 pKa = 11.49YY226 pKa = 11.83VDD228 pKa = 3.85LLYY231 pKa = 10.15WLSFGRR237 pKa = 11.84PVIIYY242 pKa = 7.24TFQPSAVAGMTDD254 pKa = 3.62DD255 pKa = 4.69ASFTIKK261 pKa = 10.71DD262 pKa = 3.92DD263 pKa = 3.65VVHH266 pKa = 6.6FSVSGGARR274 pKa = 11.84WQHH277 pKa = 5.49RR278 pKa = 11.84VWDD281 pKa = 3.99YY282 pKa = 11.73SADD285 pKa = 3.62CVYY288 pKa = 10.71VPSYY292 pKa = 10.17WNSLYY297 pKa = 10.89SYY299 pKa = 11.21VCFCEE304 pKa = 4.61SIQVSHH310 pKa = 7.37DD311 pKa = 3.21RR312 pKa = 11.84RR313 pKa = 11.84IVGLFPRR320 pKa = 11.84TKK322 pKa = 8.93MFSPMEE328 pKa = 4.2SFRR331 pKa = 11.84EE332 pKa = 3.99INPLQRR338 pKa = 11.84MRR340 pKa = 11.84YY341 pKa = 6.81TYY343 pKa = 11.21GRR345 pKa = 11.84FNVLRR350 pKa = 11.84VEE352 pKa = 4.55NKK354 pKa = 10.21GKK356 pKa = 8.8VHH358 pKa = 6.6YY359 pKa = 10.53SIGYY363 pKa = 8.0NGSAVSVDD371 pKa = 3.01INADD375 pKa = 3.39DD376 pKa = 4.69FEE378 pKa = 4.41ATRR381 pKa = 11.84IRR383 pKa = 11.84LEE385 pKa = 3.94NSKK388 pKa = 10.81KK389 pKa = 10.1PVISDD394 pKa = 3.06VEE396 pKa = 4.42RR397 pKa = 11.84ILRR400 pKa = 11.84ASSHH404 pKa = 5.66PQPVLAAAILKK415 pKa = 9.61EE416 pKa = 4.24FLDD419 pKa = 4.18KK420 pKa = 10.87TPQLPRR426 pKa = 11.84QRR428 pKa = 11.84VVSVGQPKK436 pKa = 10.25NEE438 pKa = 3.93RR439 pKa = 11.84GYY441 pKa = 11.07QSLTPLVTEE450 pKa = 4.6DD451 pKa = 3.54GKK453 pKa = 11.0NKK455 pKa = 9.8VKK457 pKa = 10.43EE458 pKa = 3.81IRR460 pKa = 11.84GAICKK465 pKa = 10.13GAVHH469 pKa = 6.61PVSSYY474 pKa = 11.55NNDD477 pKa = 2.79VRR479 pKa = 11.84CVDD482 pKa = 3.36KK483 pKa = 11.17RR484 pKa = 11.84IVAVANNTVFPQRR497 pKa = 11.84YY498 pKa = 6.95QRR500 pKa = 11.84YY501 pKa = 7.99ACAFVKK507 pKa = 10.51LLIPSDD513 pKa = 3.5QVEE516 pKa = 4.3RR517 pKa = 11.84GFPLALSDD525 pKa = 4.06VFDD528 pKa = 4.33RR529 pKa = 11.84QDD531 pKa = 3.35RR532 pKa = 11.84PSQRR536 pKa = 11.84AGFEE540 pKa = 4.16QNHH543 pKa = 4.71TTFFSTDD550 pKa = 2.79NRR552 pKa = 11.84IKK554 pKa = 10.87AFQKK558 pKa = 10.17SEE560 pKa = 3.71AYY562 pKa = 10.64ANVNDD567 pKa = 4.3PRR569 pKa = 11.84NISTTTVDD577 pKa = 2.8SRR579 pKa = 11.84VRR581 pKa = 11.84YY582 pKa = 10.06SSFTLPLAEE591 pKa = 5.2HH592 pKa = 6.36IKK594 pKa = 10.45KK595 pKa = 10.51FEE597 pKa = 3.83WYY599 pKa = 10.5AFGRR603 pKa = 11.84TPKK606 pKa = 9.72VWTEE610 pKa = 3.92MFHH613 pKa = 8.3VMASNSEE620 pKa = 4.23WILATDD626 pKa = 3.65YY627 pKa = 11.5SRR629 pKa = 11.84FDD631 pKa = 3.37GTISKK636 pKa = 10.35SFHH639 pKa = 5.66TLLRR643 pKa = 11.84LPILRR648 pKa = 11.84RR649 pKa = 11.84FFSPEE654 pKa = 3.46YY655 pKa = 9.85HH656 pKa = 6.88PEE658 pKa = 3.89VLKK661 pKa = 10.9LAEE664 pKa = 4.04QQRR667 pKa = 11.84NCPASTAHH675 pKa = 5.92GVTYY679 pKa = 10.68NSGYY683 pKa = 8.81GTVSGSADD691 pKa = 2.93TSLFNTIFNAFVAFVSICEE710 pKa = 3.93TRR712 pKa = 11.84MRR714 pKa = 11.84SLSCDD719 pKa = 2.88MAMTGKK725 pKa = 9.65PFMEE729 pKa = 4.26LVEE732 pKa = 4.45GAFEE736 pKa = 3.91EE737 pKa = 4.99LEE739 pKa = 4.19RR740 pKa = 11.84SCLFGGDD747 pKa = 4.48DD748 pKa = 6.64GILCDD753 pKa = 4.49CEE755 pKa = 4.86AGEE758 pKa = 5.01LEE760 pKa = 4.47SAAEE764 pKa = 4.19KK765 pKa = 10.2MGLKK769 pKa = 10.37LKK771 pKa = 10.63LDD773 pKa = 3.85VYY775 pKa = 10.87HH776 pKa = 6.39YY777 pKa = 11.2GPVPFLGRR785 pKa = 11.84WFVNPWVSGEE795 pKa = 4.08CMADD799 pKa = 3.27VQRR802 pKa = 11.84QVRR805 pKa = 11.84KK806 pKa = 9.91FGLTGSNLEE815 pKa = 4.07EE816 pKa = 4.41KK817 pKa = 10.35KK818 pKa = 11.04ALVAKK823 pKa = 10.51ALGYY827 pKa = 10.47LATDD831 pKa = 3.74TNTPLISHH839 pKa = 7.16WSRR842 pKa = 11.84AIIRR846 pKa = 11.84ACGDD850 pKa = 3.4LQVQPEE856 pKa = 4.42SWWARR861 pKa = 11.84MANPDD866 pKa = 3.33EE867 pKa = 5.27VVFQTSDD874 pKa = 3.1SDD876 pKa = 3.69EE877 pKa = 4.49AKK879 pKa = 10.68ALAAEE884 pKa = 4.72GCGIQVSEE892 pKa = 4.9LDD894 pKa = 4.62EE895 pKa = 4.83ICDD898 pKa = 5.85IIDD901 pKa = 3.28GWNDD905 pKa = 3.01LDD907 pKa = 4.7QFEE910 pKa = 5.47LLLDD914 pKa = 4.22LDD916 pKa = 5.05AEE918 pKa = 4.55VKK920 pKa = 10.81YY921 pKa = 10.25PVVLDD926 pKa = 3.82GSVVGEE932 pKa = 4.15VEE934 pKa = 4.2EE935 pKa = 4.58TVSITSDD942 pKa = 2.89NTSRR946 pKa = 11.84DD947 pKa = 3.46SEE949 pKa = 4.11ISSTRR954 pKa = 11.84DD955 pKa = 3.12SLKK958 pKa = 9.62TSQSAPVRR966 pKa = 11.84RR967 pKa = 11.84RR968 pKa = 11.84PSRR971 pKa = 11.84RR972 pKa = 11.84KK973 pKa = 8.69QSNPVRR979 pKa = 11.84SDD981 pKa = 3.07TTVGDD986 pKa = 3.71RR987 pKa = 11.84VVIASTKK994 pKa = 10.2VEE996 pKa = 4.2PTALGAGPAEE1006 pKa = 4.27PVNSAKK1012 pKa = 10.49LVEE1015 pKa = 4.09RR1016 pKa = 11.84AKK1018 pKa = 10.84FSSGPEE1024 pKa = 3.64
MM1 pKa = 7.82LSRR4 pKa = 11.84LYY6 pKa = 10.55HH7 pKa = 5.19VVEE10 pKa = 4.42EE11 pKa = 4.56EE12 pKa = 4.9VFEE15 pKa = 4.33PWAKK19 pKa = 10.6ALARR23 pKa = 11.84PVLHH27 pKa = 6.24VAHH30 pKa = 6.65PVVTAAYY37 pKa = 9.62DD38 pKa = 3.43VTDD41 pKa = 3.67QVSRR45 pKa = 11.84WLGPGYY51 pKa = 7.9GTCPPLSPAEE61 pKa = 3.93EE62 pKa = 3.89VVAFIRR68 pKa = 11.84SCWPDD73 pKa = 3.62LEE75 pKa = 4.57QIDD78 pKa = 4.15YY79 pKa = 11.17ALLIGCRR86 pKa = 11.84LVLLIAGAVILFNLQRR102 pKa = 11.84KK103 pKa = 6.0VLRR106 pKa = 11.84TISITNPVRR115 pKa = 11.84QYY117 pKa = 11.21VIEE120 pKa = 4.06LSKK123 pKa = 10.63IKK125 pKa = 10.23QYY127 pKa = 11.08PDD129 pKa = 3.42DD130 pKa = 3.83LRR132 pKa = 11.84KK133 pKa = 9.94KK134 pKa = 10.04FRR136 pKa = 11.84NLNVLKK142 pKa = 10.36YY143 pKa = 9.65APSLNGHH150 pKa = 5.11SHH152 pKa = 6.1GVCAAFRR159 pKa = 11.84AAADD163 pKa = 3.55DD164 pKa = 4.33TINTFIQEE172 pKa = 4.04NGFEE176 pKa = 4.34VYY178 pKa = 10.31SVGMSAKK185 pKa = 10.8DD186 pKa = 3.19MDD188 pKa = 4.82LGFEE192 pKa = 5.03GYY194 pKa = 10.49RR195 pKa = 11.84PMVEE199 pKa = 4.26DD200 pKa = 4.08KK201 pKa = 11.51DD202 pKa = 3.57MVMGFKK208 pKa = 10.51CSPVRR213 pKa = 11.84DD214 pKa = 3.49NHH216 pKa = 6.17IIKK219 pKa = 9.63MVDD222 pKa = 2.31VDD224 pKa = 4.24YY225 pKa = 11.49YY226 pKa = 11.83VDD228 pKa = 3.85LLYY231 pKa = 10.15WLSFGRR237 pKa = 11.84PVIIYY242 pKa = 7.24TFQPSAVAGMTDD254 pKa = 3.62DD255 pKa = 4.69ASFTIKK261 pKa = 10.71DD262 pKa = 3.92DD263 pKa = 3.65VVHH266 pKa = 6.6FSVSGGARR274 pKa = 11.84WQHH277 pKa = 5.49RR278 pKa = 11.84VWDD281 pKa = 3.99YY282 pKa = 11.73SADD285 pKa = 3.62CVYY288 pKa = 10.71VPSYY292 pKa = 10.17WNSLYY297 pKa = 10.89SYY299 pKa = 11.21VCFCEE304 pKa = 4.61SIQVSHH310 pKa = 7.37DD311 pKa = 3.21RR312 pKa = 11.84RR313 pKa = 11.84IVGLFPRR320 pKa = 11.84TKK322 pKa = 8.93MFSPMEE328 pKa = 4.2SFRR331 pKa = 11.84EE332 pKa = 3.99INPLQRR338 pKa = 11.84MRR340 pKa = 11.84YY341 pKa = 6.81TYY343 pKa = 11.21GRR345 pKa = 11.84FNVLRR350 pKa = 11.84VEE352 pKa = 4.55NKK354 pKa = 10.21GKK356 pKa = 8.8VHH358 pKa = 6.6YY359 pKa = 10.53SIGYY363 pKa = 8.0NGSAVSVDD371 pKa = 3.01INADD375 pKa = 3.39DD376 pKa = 4.69FEE378 pKa = 4.41ATRR381 pKa = 11.84IRR383 pKa = 11.84LEE385 pKa = 3.94NSKK388 pKa = 10.81KK389 pKa = 10.1PVISDD394 pKa = 3.06VEE396 pKa = 4.42RR397 pKa = 11.84ILRR400 pKa = 11.84ASSHH404 pKa = 5.66PQPVLAAAILKK415 pKa = 9.61EE416 pKa = 4.24FLDD419 pKa = 4.18KK420 pKa = 10.87TPQLPRR426 pKa = 11.84QRR428 pKa = 11.84VVSVGQPKK436 pKa = 10.25NEE438 pKa = 3.93RR439 pKa = 11.84GYY441 pKa = 11.07QSLTPLVTEE450 pKa = 4.6DD451 pKa = 3.54GKK453 pKa = 11.0NKK455 pKa = 9.8VKK457 pKa = 10.43EE458 pKa = 3.81IRR460 pKa = 11.84GAICKK465 pKa = 10.13GAVHH469 pKa = 6.61PVSSYY474 pKa = 11.55NNDD477 pKa = 2.79VRR479 pKa = 11.84CVDD482 pKa = 3.36KK483 pKa = 11.17RR484 pKa = 11.84IVAVANNTVFPQRR497 pKa = 11.84YY498 pKa = 6.95QRR500 pKa = 11.84YY501 pKa = 7.99ACAFVKK507 pKa = 10.51LLIPSDD513 pKa = 3.5QVEE516 pKa = 4.3RR517 pKa = 11.84GFPLALSDD525 pKa = 4.06VFDD528 pKa = 4.33RR529 pKa = 11.84QDD531 pKa = 3.35RR532 pKa = 11.84PSQRR536 pKa = 11.84AGFEE540 pKa = 4.16QNHH543 pKa = 4.71TTFFSTDD550 pKa = 2.79NRR552 pKa = 11.84IKK554 pKa = 10.87AFQKK558 pKa = 10.17SEE560 pKa = 3.71AYY562 pKa = 10.64ANVNDD567 pKa = 4.3PRR569 pKa = 11.84NISTTTVDD577 pKa = 2.8SRR579 pKa = 11.84VRR581 pKa = 11.84YY582 pKa = 10.06SSFTLPLAEE591 pKa = 5.2HH592 pKa = 6.36IKK594 pKa = 10.45KK595 pKa = 10.51FEE597 pKa = 3.83WYY599 pKa = 10.5AFGRR603 pKa = 11.84TPKK606 pKa = 9.72VWTEE610 pKa = 3.92MFHH613 pKa = 8.3VMASNSEE620 pKa = 4.23WILATDD626 pKa = 3.65YY627 pKa = 11.5SRR629 pKa = 11.84FDD631 pKa = 3.37GTISKK636 pKa = 10.35SFHH639 pKa = 5.66TLLRR643 pKa = 11.84LPILRR648 pKa = 11.84RR649 pKa = 11.84FFSPEE654 pKa = 3.46YY655 pKa = 9.85HH656 pKa = 6.88PEE658 pKa = 3.89VLKK661 pKa = 10.9LAEE664 pKa = 4.04QQRR667 pKa = 11.84NCPASTAHH675 pKa = 5.92GVTYY679 pKa = 10.68NSGYY683 pKa = 8.81GTVSGSADD691 pKa = 2.93TSLFNTIFNAFVAFVSICEE710 pKa = 3.93TRR712 pKa = 11.84MRR714 pKa = 11.84SLSCDD719 pKa = 2.88MAMTGKK725 pKa = 9.65PFMEE729 pKa = 4.26LVEE732 pKa = 4.45GAFEE736 pKa = 3.91EE737 pKa = 4.99LEE739 pKa = 4.19RR740 pKa = 11.84SCLFGGDD747 pKa = 4.48DD748 pKa = 6.64GILCDD753 pKa = 4.49CEE755 pKa = 4.86AGEE758 pKa = 5.01LEE760 pKa = 4.47SAAEE764 pKa = 4.19KK765 pKa = 10.2MGLKK769 pKa = 10.37LKK771 pKa = 10.63LDD773 pKa = 3.85VYY775 pKa = 10.87HH776 pKa = 6.39YY777 pKa = 11.2GPVPFLGRR785 pKa = 11.84WFVNPWVSGEE795 pKa = 4.08CMADD799 pKa = 3.27VQRR802 pKa = 11.84QVRR805 pKa = 11.84KK806 pKa = 9.91FGLTGSNLEE815 pKa = 4.07EE816 pKa = 4.41KK817 pKa = 10.35KK818 pKa = 11.04ALVAKK823 pKa = 10.51ALGYY827 pKa = 10.47LATDD831 pKa = 3.74TNTPLISHH839 pKa = 7.16WSRR842 pKa = 11.84AIIRR846 pKa = 11.84ACGDD850 pKa = 3.4LQVQPEE856 pKa = 4.42SWWARR861 pKa = 11.84MANPDD866 pKa = 3.33EE867 pKa = 5.27VVFQTSDD874 pKa = 3.1SDD876 pKa = 3.69EE877 pKa = 4.49AKK879 pKa = 10.68ALAAEE884 pKa = 4.72GCGIQVSEE892 pKa = 4.9LDD894 pKa = 4.62EE895 pKa = 4.83ICDD898 pKa = 5.85IIDD901 pKa = 3.28GWNDD905 pKa = 3.01LDD907 pKa = 4.7QFEE910 pKa = 5.47LLLDD914 pKa = 4.22LDD916 pKa = 5.05AEE918 pKa = 4.55VKK920 pKa = 10.81YY921 pKa = 10.25PVVLDD926 pKa = 3.82GSVVGEE932 pKa = 4.15VEE934 pKa = 4.2EE935 pKa = 4.58TVSITSDD942 pKa = 2.89NTSRR946 pKa = 11.84DD947 pKa = 3.46SEE949 pKa = 4.11ISSTRR954 pKa = 11.84DD955 pKa = 3.12SLKK958 pKa = 9.62TSQSAPVRR966 pKa = 11.84RR967 pKa = 11.84RR968 pKa = 11.84PSRR971 pKa = 11.84RR972 pKa = 11.84KK973 pKa = 8.69QSNPVRR979 pKa = 11.84SDD981 pKa = 3.07TTVGDD986 pKa = 3.71RR987 pKa = 11.84VVIASTKK994 pKa = 10.2VEE996 pKa = 4.2PTALGAGPAEE1006 pKa = 4.27PVNSAKK1012 pKa = 10.49LVEE1015 pKa = 4.09RR1016 pKa = 11.84AKK1018 pKa = 10.84FSSGPEE1024 pKa = 3.64
Molecular weight: 115.08 kDa
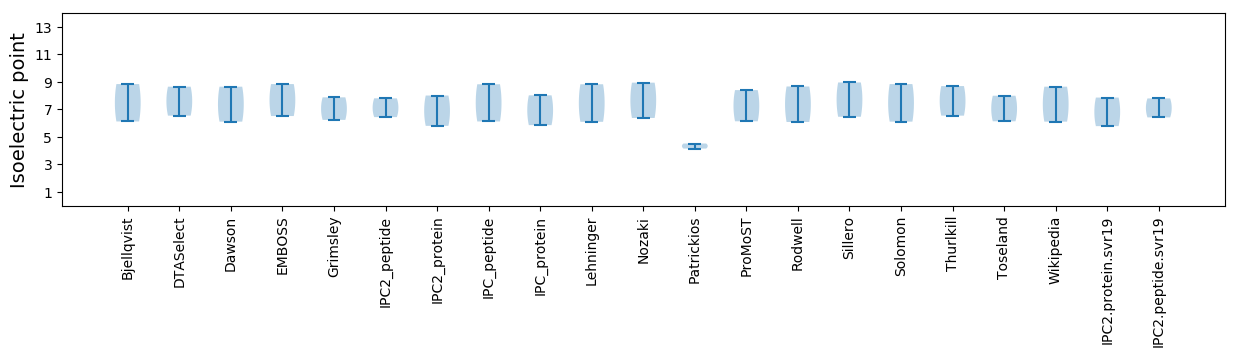
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFY0|A0A1L3KFY0_9VIRU RNA replicase OS=Beihai noda-like virus 5 OX=1922487 PE=3 SV=1
MM1 pKa = 6.86KK2 pKa = 9.17TCPVCGRR9 pKa = 11.84KK10 pKa = 9.2FKK12 pKa = 9.8TAQALRR18 pKa = 11.84QHH20 pKa = 6.8RR21 pKa = 11.84AAAHH25 pKa = 6.24RR26 pKa = 11.84GGSASKK32 pKa = 10.14PVLQGGRR39 pKa = 11.84PFPANPSTPTQQFSSEE55 pKa = 3.79NSIVLSGTDD64 pKa = 3.76RR65 pKa = 11.84LYY67 pKa = 10.6MAAVDD72 pKa = 4.92AKK74 pKa = 9.81DD75 pKa = 3.54TPLVRR80 pKa = 11.84LLIGPSLFPRR90 pKa = 11.84LAHH93 pKa = 5.75VAKK96 pKa = 9.17TFQRR100 pKa = 11.84IDD102 pKa = 3.33YY103 pKa = 10.62LMLKK107 pKa = 10.11FEE109 pKa = 4.24IVSRR113 pKa = 11.84FATITPGGYY122 pKa = 8.34VACVFTDD129 pKa = 3.91PDD131 pKa = 4.16DD132 pKa = 5.0APPNDD137 pKa = 3.92PSTLEE142 pKa = 4.89RR143 pKa = 11.84ITSTAGSVQANWWQTSTITAPPKK166 pKa = 9.38YY167 pKa = 10.06GLYY170 pKa = 9.77TSRR173 pKa = 11.84SPTSDD178 pKa = 3.3LRR180 pKa = 11.84FSSPGVFALMTEE192 pKa = 4.44IPPAQGGGFTIMCHH206 pKa = 4.38WKK208 pKa = 10.13ARR210 pKa = 11.84LYY212 pKa = 10.91HH213 pKa = 6.04PTLEE217 pKa = 4.49IDD219 pKa = 3.67VPSPFTSLIATNHH232 pKa = 4.95IKK234 pKa = 10.63AEE236 pKa = 4.1KK237 pKa = 9.12GQPRR241 pKa = 11.84YY242 pKa = 9.58AAEE245 pKa = 3.91SSEE248 pKa = 4.5DD249 pKa = 3.35NRR251 pKa = 11.84FSPEE255 pKa = 3.6VFLDD259 pKa = 3.84GLPSEE264 pKa = 5.22LADD267 pKa = 3.42GDD269 pKa = 4.09YY270 pKa = 11.5DD271 pKa = 4.55FVPEE275 pKa = 3.99TLGVFGYY282 pKa = 10.51RR283 pKa = 11.84AGTGDD288 pKa = 3.84EE289 pKa = 4.17LAAPIARR296 pKa = 11.84VRR298 pKa = 11.84LHH300 pKa = 4.99WRR302 pKa = 11.84GRR304 pKa = 11.84ANPNSNLAPWSPFSGNYY321 pKa = 9.73DD322 pKa = 3.46NPSTSDD328 pKa = 2.91NSTISNIVSWPRR340 pKa = 11.84GTRR343 pKa = 11.84FTLLQSSS350 pKa = 3.98
MM1 pKa = 6.86KK2 pKa = 9.17TCPVCGRR9 pKa = 11.84KK10 pKa = 9.2FKK12 pKa = 9.8TAQALRR18 pKa = 11.84QHH20 pKa = 6.8RR21 pKa = 11.84AAAHH25 pKa = 6.24RR26 pKa = 11.84GGSASKK32 pKa = 10.14PVLQGGRR39 pKa = 11.84PFPANPSTPTQQFSSEE55 pKa = 3.79NSIVLSGTDD64 pKa = 3.76RR65 pKa = 11.84LYY67 pKa = 10.6MAAVDD72 pKa = 4.92AKK74 pKa = 9.81DD75 pKa = 3.54TPLVRR80 pKa = 11.84LLIGPSLFPRR90 pKa = 11.84LAHH93 pKa = 5.75VAKK96 pKa = 9.17TFQRR100 pKa = 11.84IDD102 pKa = 3.33YY103 pKa = 10.62LMLKK107 pKa = 10.11FEE109 pKa = 4.24IVSRR113 pKa = 11.84FATITPGGYY122 pKa = 8.34VACVFTDD129 pKa = 3.91PDD131 pKa = 4.16DD132 pKa = 5.0APPNDD137 pKa = 3.92PSTLEE142 pKa = 4.89RR143 pKa = 11.84ITSTAGSVQANWWQTSTITAPPKK166 pKa = 9.38YY167 pKa = 10.06GLYY170 pKa = 9.77TSRR173 pKa = 11.84SPTSDD178 pKa = 3.3LRR180 pKa = 11.84FSSPGVFALMTEE192 pKa = 4.44IPPAQGGGFTIMCHH206 pKa = 4.38WKK208 pKa = 10.13ARR210 pKa = 11.84LYY212 pKa = 10.91HH213 pKa = 6.04PTLEE217 pKa = 4.49IDD219 pKa = 3.67VPSPFTSLIATNHH232 pKa = 4.95IKK234 pKa = 10.63AEE236 pKa = 4.1KK237 pKa = 9.12GQPRR241 pKa = 11.84YY242 pKa = 9.58AAEE245 pKa = 3.91SSEE248 pKa = 4.5DD249 pKa = 3.35NRR251 pKa = 11.84FSPEE255 pKa = 3.6VFLDD259 pKa = 3.84GLPSEE264 pKa = 5.22LADD267 pKa = 3.42GDD269 pKa = 4.09YY270 pKa = 11.5DD271 pKa = 4.55FVPEE275 pKa = 3.99TLGVFGYY282 pKa = 10.51RR283 pKa = 11.84AGTGDD288 pKa = 3.84EE289 pKa = 4.17LAAPIARR296 pKa = 11.84VRR298 pKa = 11.84LHH300 pKa = 4.99WRR302 pKa = 11.84GRR304 pKa = 11.84ANPNSNLAPWSPFSGNYY321 pKa = 9.73DD322 pKa = 3.46NPSTSDD328 pKa = 2.91NSTISNIVSWPRR340 pKa = 11.84GTRR343 pKa = 11.84FTLLQSSS350 pKa = 3.98
Molecular weight: 38.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1374 |
350 |
1024 |
687.0 |
76.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.933 ± 0.627 | 1.82 ± 0.351 |
6.041 ± 0.466 | 5.386 ± 1.015 |
4.876 ± 0.138 | 5.968 ± 0.609 |
1.965 ± 0.018 | 4.585 ± 0.155 |
4.294 ± 0.597 | 7.642 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.82 ± 0.203 | 3.785 ± 0.036 |
6.259 ± 1.643 | 3.202 ± 0.031 |
6.696 ± 0.064 | 8.661 ± 0.546 |
5.75 ± 1.315 | 8.297 ± 1.784 |
1.601 ± 0.059 | 3.421 ± 0.292 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
