
Paucilactobacillus wasatchensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Paucilactobacillus
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
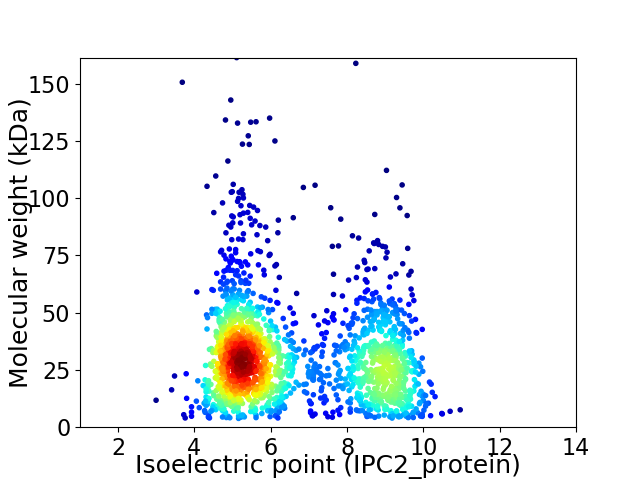
Virtual 2D-PAGE plot for 1826 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D1A8U1|A0A0D1A8U1_9LACO ATP synthase subunit beta OS=Paucilactobacillus wasatchensis OX=1335616 GN=atpD PE=3 SV=1
MM1 pKa = 7.59NKK3 pKa = 9.95RR4 pKa = 11.84LVNDD8 pKa = 3.12EE9 pKa = 4.47TEE11 pKa = 4.1IKK13 pKa = 9.86EE14 pKa = 4.06HH15 pKa = 5.78YY16 pKa = 10.39KK17 pKa = 9.7MYY19 pKa = 10.46KK20 pKa = 10.0AGGRR24 pKa = 11.84WLFASISLITFGSMLMFGTTTSKK47 pKa = 10.92AATEE51 pKa = 3.97NTEE54 pKa = 4.02ADD56 pKa = 3.97GSSSTAVTSSAAFNANDD73 pKa = 3.5STQVLSTTSNSASTEE88 pKa = 4.02TTDD91 pKa = 5.41DD92 pKa = 3.84NNSTTGDD99 pKa = 3.52AAQSEE104 pKa = 5.08VSSAAASNSTVASSSASSTYY124 pKa = 11.32SNATVATSNAASSMLLKK141 pKa = 10.08TDD143 pKa = 3.25ATVAAAATSSITMSSDD159 pKa = 2.76TVGYY163 pKa = 10.62DD164 pKa = 3.85SSNTGNGGVVNIDD177 pKa = 3.15YY178 pKa = 8.45TFTGGADD185 pKa = 3.45DD186 pKa = 4.49TFTITLPADD195 pKa = 3.36SDD197 pKa = 4.33TYY199 pKa = 11.01TLGSIPAMTTGKK211 pKa = 9.0GTMTSVKK218 pKa = 10.2NADD221 pKa = 3.28GSTTITYY228 pKa = 10.47KK229 pKa = 10.97LSTQGSYY236 pKa = 8.39TANLTLNQINNEE248 pKa = 4.16YY249 pKa = 10.03AHH251 pKa = 6.62SSAMTDD257 pKa = 2.09IGTTIKK263 pKa = 10.11TITSTINGVAQTPVTFTQIIQPSMDD288 pKa = 3.34PTGTTRR294 pKa = 11.84LHH296 pKa = 7.16PDD298 pKa = 3.01TGVVTDD304 pKa = 5.35LLPDD308 pKa = 3.28TDD310 pKa = 3.84YY311 pKa = 11.64VYY313 pKa = 10.89QFSVNEE319 pKa = 3.81ADD321 pKa = 3.88GVYY324 pKa = 10.74NDD326 pKa = 5.07GYY328 pKa = 7.96TTAKK332 pKa = 10.06VNSAANYY339 pKa = 9.73GGTTVTIPTPTGFVLDD355 pKa = 4.1SDD357 pKa = 4.85LTNQLNGYY365 pKa = 6.21TSSSGTTITQPDD377 pKa = 4.26GAGGDD382 pKa = 3.76IIISVAASDD391 pKa = 6.09GEE393 pKa = 4.47QNWNSTGSYY402 pKa = 10.24KK403 pKa = 10.69LAGYY407 pKa = 10.78YY408 pKa = 10.43DD409 pKa = 3.64VTQTDD414 pKa = 3.82EE415 pKa = 4.35EE416 pKa = 4.51QTLTAAGPATVTQVLNAAGDD436 pKa = 3.91TLTATGSGVWTDD448 pKa = 4.32SIAATDD454 pKa = 3.71TAEE457 pKa = 4.04VGAATVTASGNSSTASTQLLIDD479 pKa = 4.39GDD481 pKa = 4.28TTNDD485 pKa = 3.56PDD487 pKa = 4.26LLNSFSFTTSTAAPVDD503 pKa = 3.82DD504 pKa = 4.42AQITITIPDD513 pKa = 3.95GLDD516 pKa = 2.91ATGIKK521 pKa = 10.49VPDD524 pKa = 3.78GGANTSAYY532 pKa = 10.75LPGTTSYY539 pKa = 11.32SYY541 pKa = 10.27TLTLADD547 pKa = 4.07GSVEE551 pKa = 4.14TGTVNSGGTITPTDD565 pKa = 3.32SSAIRR570 pKa = 11.84QVVLVPNEE578 pKa = 4.08LAPGASSGTGWLSTGKK594 pKa = 9.86QVFSVYY600 pKa = 9.31GTVSATYY607 pKa = 9.82DD608 pKa = 3.63YY609 pKa = 11.36GSTIPDD615 pKa = 3.62GATLVSTISLSSEE628 pKa = 3.9DD629 pKa = 3.84ANIAEE634 pKa = 4.6MTNSVTQTVIYY645 pKa = 9.01PQDD648 pKa = 3.23AVASVGMYY656 pKa = 8.47VHH658 pKa = 6.72QISKK662 pKa = 8.16TAGYY666 pKa = 8.68EE667 pKa = 3.92NSGSISLVNTGNAGQTTNSVYY688 pKa = 10.47EE689 pKa = 4.0PIYY692 pKa = 10.8YY693 pKa = 9.96FVLPSATTVASITNLPDD710 pKa = 3.71GGVVSYY716 pKa = 7.14FTADD720 pKa = 3.38DD721 pKa = 3.62GRR723 pKa = 11.84EE724 pKa = 4.06VVEE727 pKa = 3.92VDD729 pKa = 3.39YY730 pKa = 10.47TGSGAYY736 pKa = 9.43ISTQEE741 pKa = 4.14TYY743 pKa = 10.86LIQINLTNNSDD754 pKa = 4.14ALPGTYY760 pKa = 8.56ATNLYY765 pKa = 7.97VTSPVTQLKK774 pKa = 8.19QTTPVTDD781 pKa = 3.58TSYY784 pKa = 11.62TNGDD788 pKa = 3.59ANAVSAGSGSWTISSVSTVYY808 pKa = 9.71DD809 pKa = 3.32TTLAQGNNDD818 pKa = 3.83TVPVTDD824 pKa = 4.98ASAEE828 pKa = 4.22RR829 pKa = 11.84TGDD832 pKa = 3.37TTLNFYY838 pKa = 10.9DD839 pKa = 3.65VLGNTSSATATNVSSVINLPTVGDD863 pKa = 3.89ASGSQYY869 pKa = 10.28TFEE872 pKa = 4.16LTGPIDD878 pKa = 4.19LPTSFTTSTGEE889 pKa = 4.14TEE891 pKa = 4.16LTADD895 pKa = 4.19GTPVTAQVLYY905 pKa = 9.06STSLATLNGDD915 pKa = 3.62AADD918 pKa = 3.56TSGYY922 pKa = 8.03VTADD926 pKa = 3.69QITDD930 pKa = 3.17WSAVRR935 pKa = 11.84SILIEE940 pKa = 3.91YY941 pKa = 9.96SAIPSNTSTGRR952 pKa = 11.84ISITGTSTDD961 pKa = 3.47GFEE964 pKa = 4.28GTTDD968 pKa = 3.08EE969 pKa = 4.31TGYY972 pKa = 11.35LEE974 pKa = 4.5TQFSATGYY982 pKa = 8.68DD983 pKa = 3.25TVTRR987 pKa = 11.84TGTDD991 pKa = 3.16NASLTLKK998 pKa = 9.59GTIAALSVVLNPGSLTYY1015 pKa = 10.51DD1016 pKa = 3.83GQSKK1020 pKa = 10.9ASDD1023 pKa = 3.32ATNATVTVTLADD1035 pKa = 3.53GTTQEE1040 pKa = 4.11VSISNAEE1047 pKa = 3.98GKK1049 pKa = 10.2NDD1051 pKa = 3.27FTVANDD1057 pKa = 5.05GINADD1062 pKa = 3.83SYY1064 pKa = 11.87SYY1066 pKa = 11.19ILTDD1070 pKa = 3.17SGIANVEE1077 pKa = 4.1KK1078 pKa = 10.79AVGSTIAATDD1088 pKa = 3.42VQGTIDD1094 pKa = 3.8ILSLKK1099 pKa = 9.9TSISLNDD1106 pKa = 3.53GGFTYY1111 pKa = 10.93NGDD1114 pKa = 3.85NASSASGLTASVALADD1130 pKa = 4.21GSIKK1134 pKa = 9.75TVALTSADD1142 pKa = 3.1ITVTADD1148 pKa = 3.0SSSVGSYY1155 pKa = 10.23KK1156 pKa = 10.69YY1157 pKa = 10.63SLSANGLAAVSAAVGNDD1174 pKa = 3.21YY1175 pKa = 11.0QLNNTTTTGNITITKK1190 pKa = 9.28AAYY1193 pKa = 9.51EE1194 pKa = 4.08FEE1196 pKa = 4.37LTGNEE1201 pKa = 4.18TEE1203 pKa = 4.96SYY1205 pKa = 9.73TGNQQSPDD1213 pKa = 3.3AVNYY1217 pKa = 10.66SVTLPTGEE1225 pKa = 4.52KK1226 pKa = 10.19FVLSDD1231 pKa = 3.69ADD1233 pKa = 3.49IEE1235 pKa = 4.37LVSGGTDD1242 pKa = 2.8VGSYY1246 pKa = 7.98QVQLTEE1252 pKa = 4.89LGKK1255 pKa = 10.24QVLGAIIGTNYY1266 pKa = 10.41DD1267 pKa = 3.0VSFADD1272 pKa = 3.31NATFTIIKK1280 pKa = 9.45AQAAKK1285 pKa = 10.76VSVNDD1290 pKa = 3.36QTIKK1294 pKa = 10.58EE1295 pKa = 4.31GQATPDD1301 pKa = 3.52FNVSYY1306 pKa = 11.24GDD1308 pKa = 4.13GLTSMQLTNSDD1319 pKa = 4.37FEE1321 pKa = 4.36FLGANGQILTEE1332 pKa = 4.26IPTTTGIYY1340 pKa = 8.18TVKK1343 pKa = 10.68LNAAGIAKK1351 pKa = 9.75IEE1353 pKa = 4.07AANPNYY1359 pKa = 9.95EE1360 pKa = 3.84FSEE1363 pKa = 4.59DD1364 pKa = 3.47DD1365 pKa = 4.66FIAGTYY1371 pKa = 9.31TIVSQDD1377 pKa = 3.14MANPGNPGEE1386 pKa = 4.63SGDD1389 pKa = 4.05GGLGTSEE1396 pKa = 3.99VTGTDD1401 pKa = 3.21SSDD1404 pKa = 3.38GSVTSTNSNNQVAEE1418 pKa = 4.1VTTVNKK1424 pKa = 7.8TTGSTKK1430 pKa = 10.4RR1431 pKa = 11.84LPQTGEE1437 pKa = 3.95SEE1439 pKa = 4.19NPEE1442 pKa = 4.05MAEE1445 pKa = 4.13LGLGLMSLFAMFGLLNRR1462 pKa = 11.84KK1463 pKa = 8.4KK1464 pKa = 10.45RR1465 pKa = 11.84KK1466 pKa = 7.45QQ1467 pKa = 3.19
MM1 pKa = 7.59NKK3 pKa = 9.95RR4 pKa = 11.84LVNDD8 pKa = 3.12EE9 pKa = 4.47TEE11 pKa = 4.1IKK13 pKa = 9.86EE14 pKa = 4.06HH15 pKa = 5.78YY16 pKa = 10.39KK17 pKa = 9.7MYY19 pKa = 10.46KK20 pKa = 10.0AGGRR24 pKa = 11.84WLFASISLITFGSMLMFGTTTSKK47 pKa = 10.92AATEE51 pKa = 3.97NTEE54 pKa = 4.02ADD56 pKa = 3.97GSSSTAVTSSAAFNANDD73 pKa = 3.5STQVLSTTSNSASTEE88 pKa = 4.02TTDD91 pKa = 5.41DD92 pKa = 3.84NNSTTGDD99 pKa = 3.52AAQSEE104 pKa = 5.08VSSAAASNSTVASSSASSTYY124 pKa = 11.32SNATVATSNAASSMLLKK141 pKa = 10.08TDD143 pKa = 3.25ATVAAAATSSITMSSDD159 pKa = 2.76TVGYY163 pKa = 10.62DD164 pKa = 3.85SSNTGNGGVVNIDD177 pKa = 3.15YY178 pKa = 8.45TFTGGADD185 pKa = 3.45DD186 pKa = 4.49TFTITLPADD195 pKa = 3.36SDD197 pKa = 4.33TYY199 pKa = 11.01TLGSIPAMTTGKK211 pKa = 9.0GTMTSVKK218 pKa = 10.2NADD221 pKa = 3.28GSTTITYY228 pKa = 10.47KK229 pKa = 10.97LSTQGSYY236 pKa = 8.39TANLTLNQINNEE248 pKa = 4.16YY249 pKa = 10.03AHH251 pKa = 6.62SSAMTDD257 pKa = 2.09IGTTIKK263 pKa = 10.11TITSTINGVAQTPVTFTQIIQPSMDD288 pKa = 3.34PTGTTRR294 pKa = 11.84LHH296 pKa = 7.16PDD298 pKa = 3.01TGVVTDD304 pKa = 5.35LLPDD308 pKa = 3.28TDD310 pKa = 3.84YY311 pKa = 11.64VYY313 pKa = 10.89QFSVNEE319 pKa = 3.81ADD321 pKa = 3.88GVYY324 pKa = 10.74NDD326 pKa = 5.07GYY328 pKa = 7.96TTAKK332 pKa = 10.06VNSAANYY339 pKa = 9.73GGTTVTIPTPTGFVLDD355 pKa = 4.1SDD357 pKa = 4.85LTNQLNGYY365 pKa = 6.21TSSSGTTITQPDD377 pKa = 4.26GAGGDD382 pKa = 3.76IIISVAASDD391 pKa = 6.09GEE393 pKa = 4.47QNWNSTGSYY402 pKa = 10.24KK403 pKa = 10.69LAGYY407 pKa = 10.78YY408 pKa = 10.43DD409 pKa = 3.64VTQTDD414 pKa = 3.82EE415 pKa = 4.35EE416 pKa = 4.51QTLTAAGPATVTQVLNAAGDD436 pKa = 3.91TLTATGSGVWTDD448 pKa = 4.32SIAATDD454 pKa = 3.71TAEE457 pKa = 4.04VGAATVTASGNSSTASTQLLIDD479 pKa = 4.39GDD481 pKa = 4.28TTNDD485 pKa = 3.56PDD487 pKa = 4.26LLNSFSFTTSTAAPVDD503 pKa = 3.82DD504 pKa = 4.42AQITITIPDD513 pKa = 3.95GLDD516 pKa = 2.91ATGIKK521 pKa = 10.49VPDD524 pKa = 3.78GGANTSAYY532 pKa = 10.75LPGTTSYY539 pKa = 11.32SYY541 pKa = 10.27TLTLADD547 pKa = 4.07GSVEE551 pKa = 4.14TGTVNSGGTITPTDD565 pKa = 3.32SSAIRR570 pKa = 11.84QVVLVPNEE578 pKa = 4.08LAPGASSGTGWLSTGKK594 pKa = 9.86QVFSVYY600 pKa = 9.31GTVSATYY607 pKa = 9.82DD608 pKa = 3.63YY609 pKa = 11.36GSTIPDD615 pKa = 3.62GATLVSTISLSSEE628 pKa = 3.9DD629 pKa = 3.84ANIAEE634 pKa = 4.6MTNSVTQTVIYY645 pKa = 9.01PQDD648 pKa = 3.23AVASVGMYY656 pKa = 8.47VHH658 pKa = 6.72QISKK662 pKa = 8.16TAGYY666 pKa = 8.68EE667 pKa = 3.92NSGSISLVNTGNAGQTTNSVYY688 pKa = 10.47EE689 pKa = 4.0PIYY692 pKa = 10.8YY693 pKa = 9.96FVLPSATTVASITNLPDD710 pKa = 3.71GGVVSYY716 pKa = 7.14FTADD720 pKa = 3.38DD721 pKa = 3.62GRR723 pKa = 11.84EE724 pKa = 4.06VVEE727 pKa = 3.92VDD729 pKa = 3.39YY730 pKa = 10.47TGSGAYY736 pKa = 9.43ISTQEE741 pKa = 4.14TYY743 pKa = 10.86LIQINLTNNSDD754 pKa = 4.14ALPGTYY760 pKa = 8.56ATNLYY765 pKa = 7.97VTSPVTQLKK774 pKa = 8.19QTTPVTDD781 pKa = 3.58TSYY784 pKa = 11.62TNGDD788 pKa = 3.59ANAVSAGSGSWTISSVSTVYY808 pKa = 9.71DD809 pKa = 3.32TTLAQGNNDD818 pKa = 3.83TVPVTDD824 pKa = 4.98ASAEE828 pKa = 4.22RR829 pKa = 11.84TGDD832 pKa = 3.37TTLNFYY838 pKa = 10.9DD839 pKa = 3.65VLGNTSSATATNVSSVINLPTVGDD863 pKa = 3.89ASGSQYY869 pKa = 10.28TFEE872 pKa = 4.16LTGPIDD878 pKa = 4.19LPTSFTTSTGEE889 pKa = 4.14TEE891 pKa = 4.16LTADD895 pKa = 4.19GTPVTAQVLYY905 pKa = 9.06STSLATLNGDD915 pKa = 3.62AADD918 pKa = 3.56TSGYY922 pKa = 8.03VTADD926 pKa = 3.69QITDD930 pKa = 3.17WSAVRR935 pKa = 11.84SILIEE940 pKa = 3.91YY941 pKa = 9.96SAIPSNTSTGRR952 pKa = 11.84ISITGTSTDD961 pKa = 3.47GFEE964 pKa = 4.28GTTDD968 pKa = 3.08EE969 pKa = 4.31TGYY972 pKa = 11.35LEE974 pKa = 4.5TQFSATGYY982 pKa = 8.68DD983 pKa = 3.25TVTRR987 pKa = 11.84TGTDD991 pKa = 3.16NASLTLKK998 pKa = 9.59GTIAALSVVLNPGSLTYY1015 pKa = 10.51DD1016 pKa = 3.83GQSKK1020 pKa = 10.9ASDD1023 pKa = 3.32ATNATVTVTLADD1035 pKa = 3.53GTTQEE1040 pKa = 4.11VSISNAEE1047 pKa = 3.98GKK1049 pKa = 10.2NDD1051 pKa = 3.27FTVANDD1057 pKa = 5.05GINADD1062 pKa = 3.83SYY1064 pKa = 11.87SYY1066 pKa = 11.19ILTDD1070 pKa = 3.17SGIANVEE1077 pKa = 4.1KK1078 pKa = 10.79AVGSTIAATDD1088 pKa = 3.42VQGTIDD1094 pKa = 3.8ILSLKK1099 pKa = 9.9TSISLNDD1106 pKa = 3.53GGFTYY1111 pKa = 10.93NGDD1114 pKa = 3.85NASSASGLTASVALADD1130 pKa = 4.21GSIKK1134 pKa = 9.75TVALTSADD1142 pKa = 3.1ITVTADD1148 pKa = 3.0SSSVGSYY1155 pKa = 10.23KK1156 pKa = 10.69YY1157 pKa = 10.63SLSANGLAAVSAAVGNDD1174 pKa = 3.21YY1175 pKa = 11.0QLNNTTTTGNITITKK1190 pKa = 9.28AAYY1193 pKa = 9.51EE1194 pKa = 4.08FEE1196 pKa = 4.37LTGNEE1201 pKa = 4.18TEE1203 pKa = 4.96SYY1205 pKa = 9.73TGNQQSPDD1213 pKa = 3.3AVNYY1217 pKa = 10.66SVTLPTGEE1225 pKa = 4.52KK1226 pKa = 10.19FVLSDD1231 pKa = 3.69ADD1233 pKa = 3.49IEE1235 pKa = 4.37LVSGGTDD1242 pKa = 2.8VGSYY1246 pKa = 7.98QVQLTEE1252 pKa = 4.89LGKK1255 pKa = 10.24QVLGAIIGTNYY1266 pKa = 10.41DD1267 pKa = 3.0VSFADD1272 pKa = 3.31NATFTIIKK1280 pKa = 9.45AQAAKK1285 pKa = 10.76VSVNDD1290 pKa = 3.36QTIKK1294 pKa = 10.58EE1295 pKa = 4.31GQATPDD1301 pKa = 3.52FNVSYY1306 pKa = 11.24GDD1308 pKa = 4.13GLTSMQLTNSDD1319 pKa = 4.37FEE1321 pKa = 4.36FLGANGQILTEE1332 pKa = 4.26IPTTTGIYY1340 pKa = 8.18TVKK1343 pKa = 10.68LNAAGIAKK1351 pKa = 9.75IEE1353 pKa = 4.07AANPNYY1359 pKa = 9.95EE1360 pKa = 3.84FSEE1363 pKa = 4.59DD1364 pKa = 3.47DD1365 pKa = 4.66FIAGTYY1371 pKa = 9.31TIVSQDD1377 pKa = 3.14MANPGNPGEE1386 pKa = 4.63SGDD1389 pKa = 4.05GGLGTSEE1396 pKa = 3.99VTGTDD1401 pKa = 3.21SSDD1404 pKa = 3.38GSVTSTNSNNQVAEE1418 pKa = 4.1VTTVNKK1424 pKa = 7.8TTGSTKK1430 pKa = 10.4RR1431 pKa = 11.84LPQTGEE1437 pKa = 3.95SEE1439 pKa = 4.19NPEE1442 pKa = 4.05MAEE1445 pKa = 4.13LGLGLMSLFAMFGLLNRR1462 pKa = 11.84KK1463 pKa = 8.4KK1464 pKa = 10.45RR1465 pKa = 11.84KK1466 pKa = 7.45QQ1467 pKa = 3.19
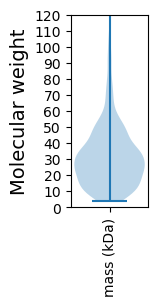
Molecular weight: 150.66 kDa
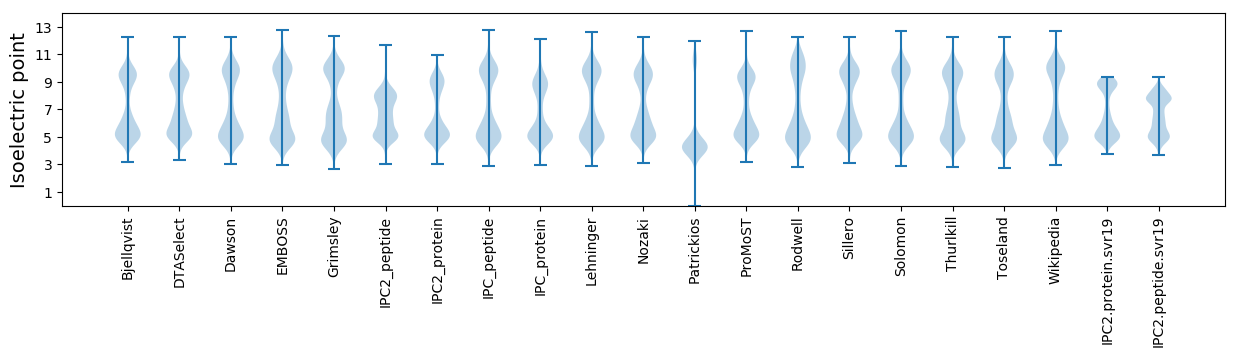
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D1A750|A0A0D1A750_9LACO Iron-sulfur cluster assembly scaffold protein OS=Paucilactobacillus wasatchensis OX=1335616 GN=iscU PE=4 SV=1
MM1 pKa = 7.2SQLDD5 pKa = 3.82NTLKK9 pKa = 11.03LLGITDD15 pKa = 3.48TNIQVFGTRR24 pKa = 11.84DD25 pKa = 3.34EE26 pKa = 4.19FHH28 pKa = 6.77GRR30 pKa = 11.84GSGRR34 pKa = 11.84KK35 pKa = 8.8KK36 pKa = 10.61YY37 pKa = 10.8LVIQAEE43 pKa = 4.2LTYY46 pKa = 9.65TLRR49 pKa = 11.84RR50 pKa = 11.84CPSCGYY56 pKa = 8.26NTLHH60 pKa = 7.05PNGHH64 pKa = 6.92KK65 pKa = 9.27LTHH68 pKa = 4.86VHH70 pKa = 6.37IAGPMDD76 pKa = 3.53RR77 pKa = 11.84PVILEE82 pKa = 4.03LNKK85 pKa = 9.92QRR87 pKa = 11.84WRR89 pKa = 11.84CSNCHH94 pKa = 5.19SACTATTPVVSTNHH108 pKa = 6.95AIGHH112 pKa = 6.07GLATHH117 pKa = 5.55VLKK120 pKa = 10.73LASKK124 pKa = 9.83SLPAKK129 pKa = 9.67TIASLTGISTNSVQRR144 pKa = 11.84ILTANIHH151 pKa = 4.5THH153 pKa = 5.71ASRR156 pKa = 11.84RR157 pKa = 11.84LPINLCFDD165 pKa = 4.42EE166 pKa = 4.97FRR168 pKa = 11.84STHH171 pKa = 5.06GSMSFICIDD180 pKa = 3.62ADD182 pKa = 3.57THH184 pKa = 6.78KK185 pKa = 10.63SVKK188 pKa = 10.16VLSDD192 pKa = 3.22RR193 pKa = 11.84LNRR196 pKa = 11.84TIKK199 pKa = 10.46QFFLSRR205 pKa = 11.84TGRR208 pKa = 11.84GSTRR212 pKa = 11.84HH213 pKa = 6.17HH214 pKa = 4.99GHH216 pKa = 6.13EE217 pKa = 4.38RR218 pKa = 11.84LLSGIRR224 pKa = 11.84ARR226 pKa = 11.84TIPP229 pKa = 3.49
MM1 pKa = 7.2SQLDD5 pKa = 3.82NTLKK9 pKa = 11.03LLGITDD15 pKa = 3.48TNIQVFGTRR24 pKa = 11.84DD25 pKa = 3.34EE26 pKa = 4.19FHH28 pKa = 6.77GRR30 pKa = 11.84GSGRR34 pKa = 11.84KK35 pKa = 8.8KK36 pKa = 10.61YY37 pKa = 10.8LVIQAEE43 pKa = 4.2LTYY46 pKa = 9.65TLRR49 pKa = 11.84RR50 pKa = 11.84CPSCGYY56 pKa = 8.26NTLHH60 pKa = 7.05PNGHH64 pKa = 6.92KK65 pKa = 9.27LTHH68 pKa = 4.86VHH70 pKa = 6.37IAGPMDD76 pKa = 3.53RR77 pKa = 11.84PVILEE82 pKa = 4.03LNKK85 pKa = 9.92QRR87 pKa = 11.84WRR89 pKa = 11.84CSNCHH94 pKa = 5.19SACTATTPVVSTNHH108 pKa = 6.95AIGHH112 pKa = 6.07GLATHH117 pKa = 5.55VLKK120 pKa = 10.73LASKK124 pKa = 9.83SLPAKK129 pKa = 9.67TIASLTGISTNSVQRR144 pKa = 11.84ILTANIHH151 pKa = 4.5THH153 pKa = 5.71ASRR156 pKa = 11.84RR157 pKa = 11.84LPINLCFDD165 pKa = 4.42EE166 pKa = 4.97FRR168 pKa = 11.84STHH171 pKa = 5.06GSMSFICIDD180 pKa = 3.62ADD182 pKa = 3.57THH184 pKa = 6.78KK185 pKa = 10.63SVKK188 pKa = 10.16VLSDD192 pKa = 3.22RR193 pKa = 11.84LNRR196 pKa = 11.84TIKK199 pKa = 10.46QFFLSRR205 pKa = 11.84TGRR208 pKa = 11.84GSTRR212 pKa = 11.84HH213 pKa = 6.17HH214 pKa = 4.99GHH216 pKa = 6.13EE217 pKa = 4.38RR218 pKa = 11.84LLSGIRR224 pKa = 11.84ARR226 pKa = 11.84TIPP229 pKa = 3.49
Molecular weight: 25.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
536271 |
37 |
1467 |
293.7 |
32.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.148 ± 0.062 | 0.454 ± 0.013 |
5.612 ± 0.051 | 5.251 ± 0.063 |
4.278 ± 0.043 | 6.702 ± 0.053 |
2.193 ± 0.024 | 7.34 ± 0.05 |
6.1 ± 0.055 | 9.783 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.706 ± 0.028 | 4.893 ± 0.041 |
3.515 ± 0.029 | 5.08 ± 0.059 |
3.89 ± 0.042 | 6.074 ± 0.067 |
6.359 ± 0.051 | 7.28 ± 0.05 |
0.982 ± 0.023 | 3.362 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
