
Phaeobacter sp. 22II1-1F12B
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Phaeobacter; unclassified Phaeobacter
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
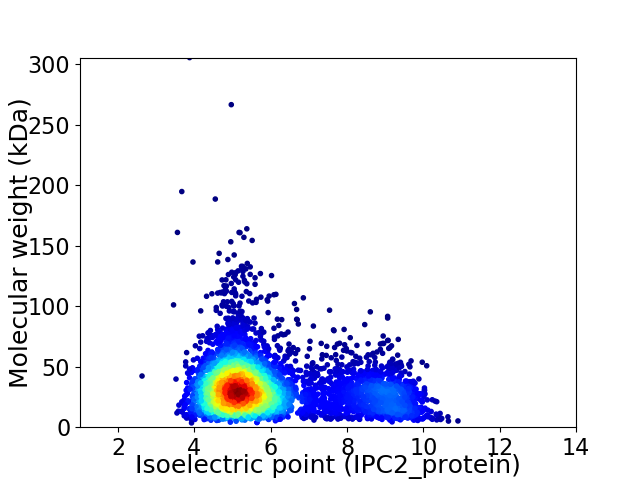
Virtual 2D-PAGE plot for 4790 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A254QXN8|A0A254QXN8_9RHOB Cytochrome P450 OS=Phaeobacter sp. 22II1-1F12B OX=1317111 GN=ATO1_00875 PE=3 SV=1
MM1 pKa = 7.49NYY3 pKa = 9.72VLEE6 pKa = 4.37TDD8 pKa = 3.41NVGVNIQDD16 pKa = 3.28IALSGDD22 pKa = 3.22FTIEE26 pKa = 3.64FNVYY30 pKa = 9.86FEE32 pKa = 5.12PGSEE36 pKa = 3.67IAKK39 pKa = 10.15NDD41 pKa = 4.38GIVSSNVDD49 pKa = 3.21GGNDD53 pKa = 3.02INFFGGIPRR62 pKa = 11.84LYY64 pKa = 9.89DD65 pKa = 3.79ANWGDD70 pKa = 3.92VVVGTSALNTGEE82 pKa = 4.03WNHH85 pKa = 5.91IAFVRR90 pKa = 11.84EE91 pKa = 3.99GGVVRR96 pKa = 11.84LYY98 pKa = 11.61VNGVNDD104 pKa = 4.68ANSTTANYY112 pKa = 9.83TDD114 pKa = 3.61VFNISTLATSVAGALEE130 pKa = 4.06GRR132 pKa = 11.84LDD134 pKa = 3.76EE135 pKa = 4.26VRR137 pKa = 11.84IWSLARR143 pKa = 11.84PGAEE147 pKa = 3.53VLANADD153 pKa = 3.34GALNLSGGVPVGLEE167 pKa = 3.43RR168 pKa = 11.84YY169 pKa = 9.52YY170 pKa = 11.13RR171 pKa = 11.84FDD173 pKa = 4.68DD174 pKa = 5.09DD175 pKa = 3.82IGHH178 pKa = 6.68IVDD181 pKa = 3.4ATGNAEE187 pKa = 4.24TGHH190 pKa = 6.91DD191 pKa = 3.87LHH193 pKa = 7.08PPTGTSIIQDD203 pKa = 3.36DD204 pKa = 4.76TVFDD208 pKa = 4.12TPVGDD213 pKa = 3.25VGGFIDD219 pKa = 3.89TKK221 pKa = 10.87VVTGLVLPTDD231 pKa = 4.15LAFLPDD237 pKa = 3.34GRR239 pKa = 11.84ALVIQKK245 pKa = 10.21SGEE248 pKa = 3.97VLLIEE253 pKa = 5.17DD254 pKa = 4.36PSVANSTKK262 pKa = 9.69TLYY265 pKa = 9.69MDD267 pKa = 5.5LSAQVEE273 pKa = 4.22NSRR276 pKa = 11.84EE277 pKa = 3.98AGLLNVVVDD286 pKa = 5.01PNFDD290 pKa = 4.07DD291 pKa = 4.07NGYY294 pKa = 10.69FYY296 pKa = 11.57LLYY299 pKa = 10.71TNLDD303 pKa = 3.61EE304 pKa = 5.18QRR306 pKa = 11.84TTVSRR311 pKa = 11.84FQHH314 pKa = 5.36QEE316 pKa = 3.41NAGGSTSTGDD326 pKa = 3.15ISSEE330 pKa = 4.12TVLWRR335 pKa = 11.84EE336 pKa = 3.34FDD338 pKa = 3.54TYY340 pKa = 10.84SEE342 pKa = 4.71KK343 pKa = 10.95DD344 pKa = 3.32HH345 pKa = 6.66QGGGLAVAYY354 pKa = 9.52EE355 pKa = 4.82PIDD358 pKa = 4.13ANDD361 pKa = 3.74PSPYY365 pKa = 10.31KK366 pKa = 10.76LYY368 pKa = 9.27ITVGEE373 pKa = 4.15EE374 pKa = 4.29FEE376 pKa = 4.66GEE378 pKa = 3.76NAADD382 pKa = 3.92LTHH385 pKa = 7.77DD386 pKa = 4.09DD387 pKa = 4.32GKK389 pKa = 10.21VHH391 pKa = 7.77RR392 pKa = 11.84INLTDD397 pKa = 3.96GSIPTDD403 pKa = 3.26NPYY406 pKa = 10.86YY407 pKa = 10.56DD408 pKa = 3.82PAVAATYY415 pKa = 8.06TPWINTQTAISTSAANTAIDD435 pKa = 3.93PEE437 pKa = 4.81GVLTTVYY444 pKa = 10.58SYY446 pKa = 11.56GLRR449 pKa = 11.84NPFRR453 pKa = 11.84AEE455 pKa = 3.65YY456 pKa = 9.84DD457 pKa = 3.72QVSGTLWIGEE467 pKa = 4.03VGGNGRR473 pKa = 11.84YY474 pKa = 9.62ASEE477 pKa = 4.76DD478 pKa = 3.14VHH480 pKa = 7.98IAYY483 pKa = 9.26PGANMGWPSEE493 pKa = 4.12EE494 pKa = 5.24GYY496 pKa = 11.08LSDD499 pKa = 5.53PNDD502 pKa = 4.24PGNPIHH508 pKa = 7.14SYY510 pKa = 9.54IHH512 pKa = 6.8ASAPGQDD519 pKa = 3.43QLPSYY524 pKa = 9.73GDD526 pKa = 3.51SGSSISGGVVYY537 pKa = 10.66RR538 pKa = 11.84GTDD541 pKa = 3.62FPEE544 pKa = 4.42EE545 pKa = 3.68FQGAYY550 pKa = 10.14FYY552 pKa = 11.49GDD554 pKa = 3.16WVRR557 pKa = 11.84NWIRR561 pKa = 11.84YY562 pKa = 9.4LEE564 pKa = 4.05VDD566 pKa = 3.77YY567 pKa = 11.6SSGEE571 pKa = 3.99PVVVSDD577 pKa = 4.04NHH579 pKa = 6.25FKK581 pKa = 10.96NATGQVLTFAEE592 pKa = 4.71APDD595 pKa = 3.5GTLYY599 pKa = 11.18YY600 pKa = 9.49ITTFQTGNIFNFEE613 pKa = 3.95GAVNRR618 pKa = 11.84LEE620 pKa = 4.01YY621 pKa = 10.8SSTNTAPAGTGIILDD636 pKa = 4.16PGEE639 pKa = 4.41EE640 pKa = 4.05ASLTAPHH647 pKa = 6.45TVTFEE652 pKa = 3.91TDD654 pKa = 2.92AFDD657 pKa = 5.13PDD659 pKa = 3.82GDD661 pKa = 4.68AITYY665 pKa = 7.36EE666 pKa = 4.18WSFGDD671 pKa = 4.97GPDD674 pKa = 3.74IDD676 pKa = 4.88GDD678 pKa = 4.48GIGDD682 pKa = 4.03GATSTEE688 pKa = 4.08MNPTYY693 pKa = 10.21TYY695 pKa = 10.21TSEE698 pKa = 3.99GQYY701 pKa = 9.53TVEE704 pKa = 5.79LIVTDD709 pKa = 4.01ANGAQTVYY717 pKa = 9.7NPKK720 pKa = 10.37VITVGEE726 pKa = 4.19APVVTINTPTQGQTFRR742 pKa = 11.84AGEE745 pKa = 4.17TVIFTGSAFDD755 pKa = 4.47ANDD758 pKa = 3.66GVLTGEE764 pKa = 4.14DD765 pKa = 3.85VFWSVNLAHH774 pKa = 6.7NEE776 pKa = 4.05HH777 pKa = 6.31YY778 pKa = 10.4HH779 pKa = 6.21PEE781 pKa = 3.92VSAVEE786 pKa = 4.02NTPGGFSFVVPTDD799 pKa = 2.97GHH801 pKa = 6.92DD802 pKa = 3.32YY803 pKa = 10.49YY804 pKa = 11.26EE805 pKa = 4.29NVGYY809 pKa = 9.91QVSLTAIDD817 pKa = 3.75SSGLRR822 pKa = 11.84TTQSVTIFPEE832 pKa = 4.33EE833 pKa = 4.41SITTYY838 pKa = 11.48DD839 pKa = 3.57MPDD842 pKa = 2.76VSGFNFFIDD851 pKa = 4.97GIGYY855 pKa = 10.11SGDD858 pKa = 3.36QVRR861 pKa = 11.84DD862 pKa = 3.58NIIGFEE868 pKa = 4.18HH869 pKa = 6.05TVSVQASYY877 pKa = 11.21IDD879 pKa = 5.35AGFLWEE885 pKa = 4.43FSHH888 pKa = 7.29WEE890 pKa = 4.22DD891 pKa = 5.15DD892 pKa = 3.84PSLTSTSRR900 pKa = 11.84QFIVPEE906 pKa = 4.0TDD908 pKa = 3.04SVLRR912 pKa = 11.84PVYY915 pKa = 10.52AQTDD919 pKa = 3.67VVGPVNAEE927 pKa = 3.7DD928 pKa = 4.63DD929 pKa = 3.92VFARR933 pKa = 11.84DD934 pKa = 3.55LASLNGNQIQTIGNVLLNDD953 pKa = 4.73DD954 pKa = 5.05NPTGDD959 pKa = 3.57PFSVVGLNGYY969 pKa = 9.46KK970 pKa = 8.97VTGAPKK976 pKa = 10.01TDD978 pKa = 3.51DD979 pKa = 4.49TIDD982 pKa = 3.53SNNKK986 pKa = 9.37YY987 pKa = 9.77GWIWGDD993 pKa = 2.82QGGQFRR999 pKa = 11.84VNANGTVEE1007 pKa = 4.3FRR1009 pKa = 11.84DD1010 pKa = 3.65RR1011 pKa = 11.84DD1012 pKa = 3.69LQFAGLTAGQSITTTINYY1030 pKa = 9.57LIADD1034 pKa = 4.03GDD1036 pKa = 4.11DD1037 pKa = 3.38TDD1039 pKa = 4.07EE1040 pKa = 5.08ASLSLTINGAGTANTAPTGAGILLDD1065 pKa = 4.27PGEE1068 pKa = 5.16DD1069 pKa = 3.46SSASAPHH1076 pKa = 6.16TVTFDD1081 pKa = 3.38SDD1083 pKa = 3.55VTDD1086 pKa = 4.31AEE1088 pKa = 4.51GHH1090 pKa = 4.92TLSYY1094 pKa = 10.9LWDD1097 pKa = 4.63FGDD1100 pKa = 4.09GTTSTLAAPEE1110 pKa = 4.05HH1111 pKa = 6.35TYY1113 pKa = 11.2SVVGFYY1119 pKa = 9.63TVSLTVTDD1127 pKa = 3.8EE1128 pKa = 4.45LGAEE1132 pKa = 4.29TVFDD1136 pKa = 4.14EE1137 pKa = 4.98VSIDD1141 pKa = 3.59VGNAPPVAVADD1152 pKa = 4.07SFTLPVGFDD1161 pKa = 3.25PMANQLNILDD1171 pKa = 4.44NDD1173 pKa = 3.84YY1174 pKa = 11.65DD1175 pKa = 4.41PDD1177 pKa = 4.46GSLNMQIVEE1186 pKa = 4.31IVSGPSFGTVEE1197 pKa = 4.2ILDD1200 pKa = 4.17AAQEE1204 pKa = 4.02LSDD1207 pKa = 3.95RR1208 pKa = 11.84GLPAGQYY1215 pKa = 9.6GQAEE1219 pKa = 4.43YY1220 pKa = 9.22TAFDD1224 pKa = 4.6PNFEE1228 pKa = 4.29GTDD1231 pKa = 3.23SFTYY1235 pKa = 10.08RR1236 pKa = 11.84VQDD1239 pKa = 4.09NEE1241 pKa = 4.42GMWSNIVTAEE1251 pKa = 3.85IVVQNAPVLTLDD1263 pKa = 4.81AVDD1266 pKa = 4.66DD1267 pKa = 5.07GYY1269 pKa = 11.97AVLVADD1275 pKa = 4.48IASGSWVQIQGNIFDD1290 pKa = 5.09NDD1292 pKa = 3.72TSDD1295 pKa = 3.45GALNLLQLDD1304 pKa = 3.92GFKK1307 pKa = 10.9PFDD1310 pKa = 3.86VPDD1313 pKa = 3.63STTSFDD1319 pKa = 3.37SQGRR1323 pKa = 11.84LKK1325 pKa = 10.26IRR1327 pKa = 11.84ADD1329 pKa = 2.97NGGFFRR1335 pKa = 11.84IGSDD1339 pKa = 3.27GSIEE1343 pKa = 4.15FKK1345 pKa = 11.03DD1346 pKa = 3.45RR1347 pKa = 11.84DD1348 pKa = 3.5FDD1350 pKa = 4.03FTGLSSSEE1358 pKa = 4.27SAVTTISYY1366 pKa = 8.85TIGDD1370 pKa = 4.0GTDD1373 pKa = 2.78TDD1375 pKa = 3.91AAFISLTINGSGNVNTAPTGAGILLDD1401 pKa = 4.45PGEE1404 pKa = 5.24DD1405 pKa = 3.5SATSAPHH1412 pKa = 5.85TVTFDD1417 pKa = 3.38SDD1419 pKa = 3.55VTDD1422 pKa = 4.31AEE1424 pKa = 4.51GHH1426 pKa = 4.92TLSYY1430 pKa = 10.9LWDD1433 pKa = 4.63FGDD1436 pKa = 4.09GTTSTLAAPEE1446 pKa = 4.01HH1447 pKa = 6.5TYY1449 pKa = 11.53NSIGFYY1455 pKa = 10.33NVSLTVTDD1463 pKa = 3.91EE1464 pKa = 4.29LGAATVFNEE1473 pKa = 3.69VSIDD1477 pKa = 3.6VGNAPPVAVADD1488 pKa = 4.07SFTLPVGFDD1497 pKa = 3.25PVANHH1502 pKa = 6.61LHH1504 pKa = 6.47ILDD1507 pKa = 4.24NDD1509 pKa = 3.77YY1510 pKa = 11.66DD1511 pKa = 4.51PDD1513 pKa = 4.43GSLNMQVVEE1522 pKa = 4.4IVSGPSFGTVEE1533 pKa = 4.15MMDD1536 pKa = 3.89TAQEE1540 pKa = 3.95LADD1543 pKa = 4.09RR1544 pKa = 11.84GLPAGHH1550 pKa = 6.59YY1551 pKa = 9.77GHH1553 pKa = 7.49AEE1555 pKa = 3.93YY1556 pKa = 8.51ATFDD1560 pKa = 4.19PNFEE1564 pKa = 4.26GTDD1567 pKa = 3.23SFTYY1571 pKa = 10.08RR1572 pKa = 11.84VQDD1575 pKa = 4.9NEE1577 pKa = 4.61GTWSNIVTAEE1587 pKa = 4.14INVQSAPSTPGGALASVNSGSGLDD1611 pKa = 3.42IQDD1614 pKa = 3.13ISVAGDD1620 pKa = 3.16FTVEE1624 pKa = 3.61FWIAFNPGTNTNKK1637 pKa = 9.98YY1638 pKa = 10.18DD1639 pKa = 3.35SAFAGGVYY1647 pKa = 10.36DD1648 pKa = 5.06DD1649 pKa = 5.07TPGDD1653 pKa = 3.94DD1654 pKa = 3.91PSTDD1658 pKa = 3.15GVFGNDD1664 pKa = 3.24FNFHH1668 pKa = 6.56AGLARR1673 pKa = 11.84LYY1675 pKa = 9.66STEE1678 pKa = 4.03IPSGGDD1684 pKa = 3.08AIVANTALTSNGVANHH1700 pKa = 5.81VALVRR1705 pKa = 11.84EE1706 pKa = 4.52GNNFSIYY1713 pKa = 9.8MNGVLDD1719 pKa = 4.0STSTLALDD1727 pKa = 3.78GDD1729 pKa = 4.35LLIEE1733 pKa = 4.15RR1734 pKa = 11.84LATNVRR1740 pKa = 11.84ATKK1743 pKa = 10.38ALSGTLDD1750 pKa = 3.61EE1751 pKa = 5.02FRR1753 pKa = 11.84VWDD1756 pKa = 4.08DD1757 pKa = 3.25ARR1759 pKa = 11.84SASEE1763 pKa = 3.29IATYY1767 pKa = 9.84MNQEE1771 pKa = 4.12VASNADD1777 pKa = 3.34DD1778 pKa = 4.02LLRR1781 pKa = 11.84YY1782 pKa = 9.17YY1783 pKa = 10.78QFDD1786 pKa = 4.42DD1787 pKa = 3.59DD1788 pKa = 4.58TFVIDD1793 pKa = 3.44STGNAEE1799 pKa = 4.05AAGDD1803 pKa = 3.85LSYY1806 pKa = 11.72ASISGVDD1813 pKa = 3.03WFEE1816 pKa = 4.01FSDD1819 pKa = 3.46WLGAA1823 pKa = 3.78
MM1 pKa = 7.49NYY3 pKa = 9.72VLEE6 pKa = 4.37TDD8 pKa = 3.41NVGVNIQDD16 pKa = 3.28IALSGDD22 pKa = 3.22FTIEE26 pKa = 3.64FNVYY30 pKa = 9.86FEE32 pKa = 5.12PGSEE36 pKa = 3.67IAKK39 pKa = 10.15NDD41 pKa = 4.38GIVSSNVDD49 pKa = 3.21GGNDD53 pKa = 3.02INFFGGIPRR62 pKa = 11.84LYY64 pKa = 9.89DD65 pKa = 3.79ANWGDD70 pKa = 3.92VVVGTSALNTGEE82 pKa = 4.03WNHH85 pKa = 5.91IAFVRR90 pKa = 11.84EE91 pKa = 3.99GGVVRR96 pKa = 11.84LYY98 pKa = 11.61VNGVNDD104 pKa = 4.68ANSTTANYY112 pKa = 9.83TDD114 pKa = 3.61VFNISTLATSVAGALEE130 pKa = 4.06GRR132 pKa = 11.84LDD134 pKa = 3.76EE135 pKa = 4.26VRR137 pKa = 11.84IWSLARR143 pKa = 11.84PGAEE147 pKa = 3.53VLANADD153 pKa = 3.34GALNLSGGVPVGLEE167 pKa = 3.43RR168 pKa = 11.84YY169 pKa = 9.52YY170 pKa = 11.13RR171 pKa = 11.84FDD173 pKa = 4.68DD174 pKa = 5.09DD175 pKa = 3.82IGHH178 pKa = 6.68IVDD181 pKa = 3.4ATGNAEE187 pKa = 4.24TGHH190 pKa = 6.91DD191 pKa = 3.87LHH193 pKa = 7.08PPTGTSIIQDD203 pKa = 3.36DD204 pKa = 4.76TVFDD208 pKa = 4.12TPVGDD213 pKa = 3.25VGGFIDD219 pKa = 3.89TKK221 pKa = 10.87VVTGLVLPTDD231 pKa = 4.15LAFLPDD237 pKa = 3.34GRR239 pKa = 11.84ALVIQKK245 pKa = 10.21SGEE248 pKa = 3.97VLLIEE253 pKa = 5.17DD254 pKa = 4.36PSVANSTKK262 pKa = 9.69TLYY265 pKa = 9.69MDD267 pKa = 5.5LSAQVEE273 pKa = 4.22NSRR276 pKa = 11.84EE277 pKa = 3.98AGLLNVVVDD286 pKa = 5.01PNFDD290 pKa = 4.07DD291 pKa = 4.07NGYY294 pKa = 10.69FYY296 pKa = 11.57LLYY299 pKa = 10.71TNLDD303 pKa = 3.61EE304 pKa = 5.18QRR306 pKa = 11.84TTVSRR311 pKa = 11.84FQHH314 pKa = 5.36QEE316 pKa = 3.41NAGGSTSTGDD326 pKa = 3.15ISSEE330 pKa = 4.12TVLWRR335 pKa = 11.84EE336 pKa = 3.34FDD338 pKa = 3.54TYY340 pKa = 10.84SEE342 pKa = 4.71KK343 pKa = 10.95DD344 pKa = 3.32HH345 pKa = 6.66QGGGLAVAYY354 pKa = 9.52EE355 pKa = 4.82PIDD358 pKa = 4.13ANDD361 pKa = 3.74PSPYY365 pKa = 10.31KK366 pKa = 10.76LYY368 pKa = 9.27ITVGEE373 pKa = 4.15EE374 pKa = 4.29FEE376 pKa = 4.66GEE378 pKa = 3.76NAADD382 pKa = 3.92LTHH385 pKa = 7.77DD386 pKa = 4.09DD387 pKa = 4.32GKK389 pKa = 10.21VHH391 pKa = 7.77RR392 pKa = 11.84INLTDD397 pKa = 3.96GSIPTDD403 pKa = 3.26NPYY406 pKa = 10.86YY407 pKa = 10.56DD408 pKa = 3.82PAVAATYY415 pKa = 8.06TPWINTQTAISTSAANTAIDD435 pKa = 3.93PEE437 pKa = 4.81GVLTTVYY444 pKa = 10.58SYY446 pKa = 11.56GLRR449 pKa = 11.84NPFRR453 pKa = 11.84AEE455 pKa = 3.65YY456 pKa = 9.84DD457 pKa = 3.72QVSGTLWIGEE467 pKa = 4.03VGGNGRR473 pKa = 11.84YY474 pKa = 9.62ASEE477 pKa = 4.76DD478 pKa = 3.14VHH480 pKa = 7.98IAYY483 pKa = 9.26PGANMGWPSEE493 pKa = 4.12EE494 pKa = 5.24GYY496 pKa = 11.08LSDD499 pKa = 5.53PNDD502 pKa = 4.24PGNPIHH508 pKa = 7.14SYY510 pKa = 9.54IHH512 pKa = 6.8ASAPGQDD519 pKa = 3.43QLPSYY524 pKa = 9.73GDD526 pKa = 3.51SGSSISGGVVYY537 pKa = 10.66RR538 pKa = 11.84GTDD541 pKa = 3.62FPEE544 pKa = 4.42EE545 pKa = 3.68FQGAYY550 pKa = 10.14FYY552 pKa = 11.49GDD554 pKa = 3.16WVRR557 pKa = 11.84NWIRR561 pKa = 11.84YY562 pKa = 9.4LEE564 pKa = 4.05VDD566 pKa = 3.77YY567 pKa = 11.6SSGEE571 pKa = 3.99PVVVSDD577 pKa = 4.04NHH579 pKa = 6.25FKK581 pKa = 10.96NATGQVLTFAEE592 pKa = 4.71APDD595 pKa = 3.5GTLYY599 pKa = 11.18YY600 pKa = 9.49ITTFQTGNIFNFEE613 pKa = 3.95GAVNRR618 pKa = 11.84LEE620 pKa = 4.01YY621 pKa = 10.8SSTNTAPAGTGIILDD636 pKa = 4.16PGEE639 pKa = 4.41EE640 pKa = 4.05ASLTAPHH647 pKa = 6.45TVTFEE652 pKa = 3.91TDD654 pKa = 2.92AFDD657 pKa = 5.13PDD659 pKa = 3.82GDD661 pKa = 4.68AITYY665 pKa = 7.36EE666 pKa = 4.18WSFGDD671 pKa = 4.97GPDD674 pKa = 3.74IDD676 pKa = 4.88GDD678 pKa = 4.48GIGDD682 pKa = 4.03GATSTEE688 pKa = 4.08MNPTYY693 pKa = 10.21TYY695 pKa = 10.21TSEE698 pKa = 3.99GQYY701 pKa = 9.53TVEE704 pKa = 5.79LIVTDD709 pKa = 4.01ANGAQTVYY717 pKa = 9.7NPKK720 pKa = 10.37VITVGEE726 pKa = 4.19APVVTINTPTQGQTFRR742 pKa = 11.84AGEE745 pKa = 4.17TVIFTGSAFDD755 pKa = 4.47ANDD758 pKa = 3.66GVLTGEE764 pKa = 4.14DD765 pKa = 3.85VFWSVNLAHH774 pKa = 6.7NEE776 pKa = 4.05HH777 pKa = 6.31YY778 pKa = 10.4HH779 pKa = 6.21PEE781 pKa = 3.92VSAVEE786 pKa = 4.02NTPGGFSFVVPTDD799 pKa = 2.97GHH801 pKa = 6.92DD802 pKa = 3.32YY803 pKa = 10.49YY804 pKa = 11.26EE805 pKa = 4.29NVGYY809 pKa = 9.91QVSLTAIDD817 pKa = 3.75SSGLRR822 pKa = 11.84TTQSVTIFPEE832 pKa = 4.33EE833 pKa = 4.41SITTYY838 pKa = 11.48DD839 pKa = 3.57MPDD842 pKa = 2.76VSGFNFFIDD851 pKa = 4.97GIGYY855 pKa = 10.11SGDD858 pKa = 3.36QVRR861 pKa = 11.84DD862 pKa = 3.58NIIGFEE868 pKa = 4.18HH869 pKa = 6.05TVSVQASYY877 pKa = 11.21IDD879 pKa = 5.35AGFLWEE885 pKa = 4.43FSHH888 pKa = 7.29WEE890 pKa = 4.22DD891 pKa = 5.15DD892 pKa = 3.84PSLTSTSRR900 pKa = 11.84QFIVPEE906 pKa = 4.0TDD908 pKa = 3.04SVLRR912 pKa = 11.84PVYY915 pKa = 10.52AQTDD919 pKa = 3.67VVGPVNAEE927 pKa = 3.7DD928 pKa = 4.63DD929 pKa = 3.92VFARR933 pKa = 11.84DD934 pKa = 3.55LASLNGNQIQTIGNVLLNDD953 pKa = 4.73DD954 pKa = 5.05NPTGDD959 pKa = 3.57PFSVVGLNGYY969 pKa = 9.46KK970 pKa = 8.97VTGAPKK976 pKa = 10.01TDD978 pKa = 3.51DD979 pKa = 4.49TIDD982 pKa = 3.53SNNKK986 pKa = 9.37YY987 pKa = 9.77GWIWGDD993 pKa = 2.82QGGQFRR999 pKa = 11.84VNANGTVEE1007 pKa = 4.3FRR1009 pKa = 11.84DD1010 pKa = 3.65RR1011 pKa = 11.84DD1012 pKa = 3.69LQFAGLTAGQSITTTINYY1030 pKa = 9.57LIADD1034 pKa = 4.03GDD1036 pKa = 4.11DD1037 pKa = 3.38TDD1039 pKa = 4.07EE1040 pKa = 5.08ASLSLTINGAGTANTAPTGAGILLDD1065 pKa = 4.27PGEE1068 pKa = 5.16DD1069 pKa = 3.46SSASAPHH1076 pKa = 6.16TVTFDD1081 pKa = 3.38SDD1083 pKa = 3.55VTDD1086 pKa = 4.31AEE1088 pKa = 4.51GHH1090 pKa = 4.92TLSYY1094 pKa = 10.9LWDD1097 pKa = 4.63FGDD1100 pKa = 4.09GTTSTLAAPEE1110 pKa = 4.05HH1111 pKa = 6.35TYY1113 pKa = 11.2SVVGFYY1119 pKa = 9.63TVSLTVTDD1127 pKa = 3.8EE1128 pKa = 4.45LGAEE1132 pKa = 4.29TVFDD1136 pKa = 4.14EE1137 pKa = 4.98VSIDD1141 pKa = 3.59VGNAPPVAVADD1152 pKa = 4.07SFTLPVGFDD1161 pKa = 3.25PMANQLNILDD1171 pKa = 4.44NDD1173 pKa = 3.84YY1174 pKa = 11.65DD1175 pKa = 4.41PDD1177 pKa = 4.46GSLNMQIVEE1186 pKa = 4.31IVSGPSFGTVEE1197 pKa = 4.2ILDD1200 pKa = 4.17AAQEE1204 pKa = 4.02LSDD1207 pKa = 3.95RR1208 pKa = 11.84GLPAGQYY1215 pKa = 9.6GQAEE1219 pKa = 4.43YY1220 pKa = 9.22TAFDD1224 pKa = 4.6PNFEE1228 pKa = 4.29GTDD1231 pKa = 3.23SFTYY1235 pKa = 10.08RR1236 pKa = 11.84VQDD1239 pKa = 4.09NEE1241 pKa = 4.42GMWSNIVTAEE1251 pKa = 3.85IVVQNAPVLTLDD1263 pKa = 4.81AVDD1266 pKa = 4.66DD1267 pKa = 5.07GYY1269 pKa = 11.97AVLVADD1275 pKa = 4.48IASGSWVQIQGNIFDD1290 pKa = 5.09NDD1292 pKa = 3.72TSDD1295 pKa = 3.45GALNLLQLDD1304 pKa = 3.92GFKK1307 pKa = 10.9PFDD1310 pKa = 3.86VPDD1313 pKa = 3.63STTSFDD1319 pKa = 3.37SQGRR1323 pKa = 11.84LKK1325 pKa = 10.26IRR1327 pKa = 11.84ADD1329 pKa = 2.97NGGFFRR1335 pKa = 11.84IGSDD1339 pKa = 3.27GSIEE1343 pKa = 4.15FKK1345 pKa = 11.03DD1346 pKa = 3.45RR1347 pKa = 11.84DD1348 pKa = 3.5FDD1350 pKa = 4.03FTGLSSSEE1358 pKa = 4.27SAVTTISYY1366 pKa = 8.85TIGDD1370 pKa = 4.0GTDD1373 pKa = 2.78TDD1375 pKa = 3.91AAFISLTINGSGNVNTAPTGAGILLDD1401 pKa = 4.45PGEE1404 pKa = 5.24DD1405 pKa = 3.5SATSAPHH1412 pKa = 5.85TVTFDD1417 pKa = 3.38SDD1419 pKa = 3.55VTDD1422 pKa = 4.31AEE1424 pKa = 4.51GHH1426 pKa = 4.92TLSYY1430 pKa = 10.9LWDD1433 pKa = 4.63FGDD1436 pKa = 4.09GTTSTLAAPEE1446 pKa = 4.01HH1447 pKa = 6.5TYY1449 pKa = 11.53NSIGFYY1455 pKa = 10.33NVSLTVTDD1463 pKa = 3.91EE1464 pKa = 4.29LGAATVFNEE1473 pKa = 3.69VSIDD1477 pKa = 3.6VGNAPPVAVADD1488 pKa = 4.07SFTLPVGFDD1497 pKa = 3.25PVANHH1502 pKa = 6.61LHH1504 pKa = 6.47ILDD1507 pKa = 4.24NDD1509 pKa = 3.77YY1510 pKa = 11.66DD1511 pKa = 4.51PDD1513 pKa = 4.43GSLNMQVVEE1522 pKa = 4.4IVSGPSFGTVEE1533 pKa = 4.15MMDD1536 pKa = 3.89TAQEE1540 pKa = 3.95LADD1543 pKa = 4.09RR1544 pKa = 11.84GLPAGHH1550 pKa = 6.59YY1551 pKa = 9.77GHH1553 pKa = 7.49AEE1555 pKa = 3.93YY1556 pKa = 8.51ATFDD1560 pKa = 4.19PNFEE1564 pKa = 4.26GTDD1567 pKa = 3.23SFTYY1571 pKa = 10.08RR1572 pKa = 11.84VQDD1575 pKa = 4.9NEE1577 pKa = 4.61GTWSNIVTAEE1587 pKa = 4.14INVQSAPSTPGGALASVNSGSGLDD1611 pKa = 3.42IQDD1614 pKa = 3.13ISVAGDD1620 pKa = 3.16FTVEE1624 pKa = 3.61FWIAFNPGTNTNKK1637 pKa = 9.98YY1638 pKa = 10.18DD1639 pKa = 3.35SAFAGGVYY1647 pKa = 10.36DD1648 pKa = 5.06DD1649 pKa = 5.07TPGDD1653 pKa = 3.94DD1654 pKa = 3.91PSTDD1658 pKa = 3.15GVFGNDD1664 pKa = 3.24FNFHH1668 pKa = 6.56AGLARR1673 pKa = 11.84LYY1675 pKa = 9.66STEE1678 pKa = 4.03IPSGGDD1684 pKa = 3.08AIVANTALTSNGVANHH1700 pKa = 5.81VALVRR1705 pKa = 11.84EE1706 pKa = 4.52GNNFSIYY1713 pKa = 9.8MNGVLDD1719 pKa = 4.0STSTLALDD1727 pKa = 3.78GDD1729 pKa = 4.35LLIEE1733 pKa = 4.15RR1734 pKa = 11.84LATNVRR1740 pKa = 11.84ATKK1743 pKa = 10.38ALSGTLDD1750 pKa = 3.61EE1751 pKa = 5.02FRR1753 pKa = 11.84VWDD1756 pKa = 4.08DD1757 pKa = 3.25ARR1759 pKa = 11.84SASEE1763 pKa = 3.29IATYY1767 pKa = 9.84MNQEE1771 pKa = 4.12VASNADD1777 pKa = 3.34DD1778 pKa = 4.02LLRR1781 pKa = 11.84YY1782 pKa = 9.17YY1783 pKa = 10.78QFDD1786 pKa = 4.42DD1787 pKa = 3.59DD1788 pKa = 4.58TFVIDD1793 pKa = 3.44STGNAEE1799 pKa = 4.05AAGDD1803 pKa = 3.85LSYY1806 pKa = 11.72ASISGVDD1813 pKa = 3.03WFEE1816 pKa = 4.01FSDD1819 pKa = 3.46WLGAA1823 pKa = 3.78
Molecular weight: 194.8 kDa
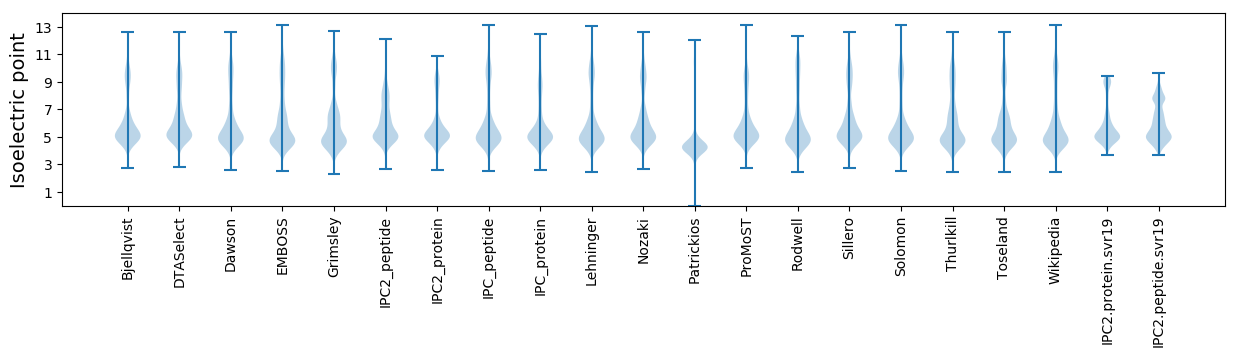
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A254QLH7|A0A254QLH7_9RHOB Sorbitol dehydrogenase OS=Phaeobacter sp. 22II1-1F12B OX=1317111 GN=ATO1_20225 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.43EE41 pKa = 3.72LSAA44 pKa = 5.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.43EE41 pKa = 3.72LSAA44 pKa = 5.03
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1505669 |
31 |
2868 |
314.3 |
34.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.513 ± 0.05 | 0.899 ± 0.012 |
6.084 ± 0.034 | 6.294 ± 0.035 |
3.795 ± 0.023 | 8.573 ± 0.036 |
2.012 ± 0.02 | 5.365 ± 0.026 |
3.342 ± 0.026 | 9.968 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.887 ± 0.017 | 2.747 ± 0.017 |
5.012 ± 0.024 | 3.181 ± 0.018 |
6.551 ± 0.036 | 5.529 ± 0.022 |
5.378 ± 0.02 | 7.166 ± 0.028 |
1.392 ± 0.015 | 2.312 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
