
Actinomyces sp. oral taxon 175 str. F0384
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Actinomyces; unclassified Actinomyces; Actinomyces sp. oral taxon 175
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
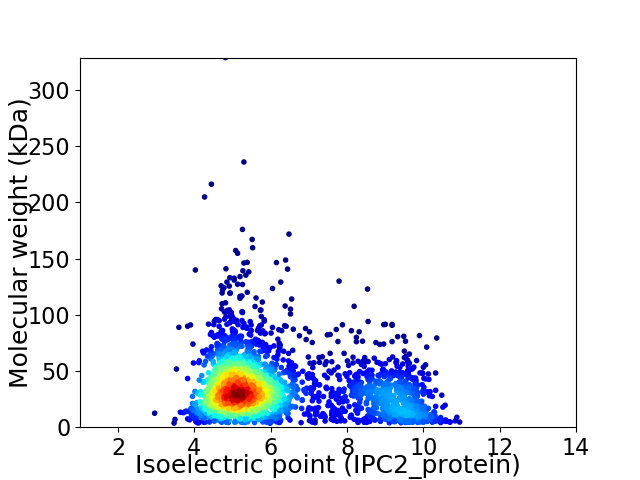
Virtual 2D-PAGE plot for 2638 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
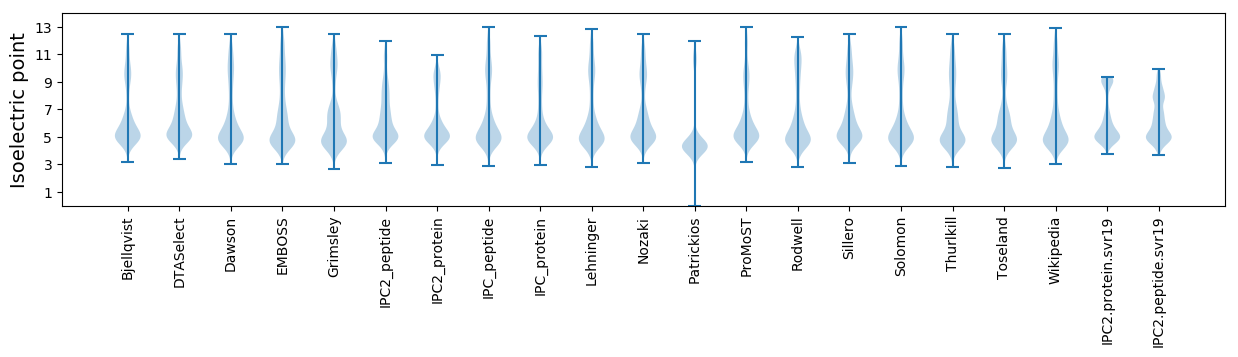
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F9PJC1|F9PJC1_9ACTO Transporter major facilitator family protein OS=Actinomyces sp. oral taxon 175 str. F0384 OX=944560 GN=HMPREF9058_2037 PE=4 SV=1
MM1 pKa = 7.03AQSVHH6 pKa = 4.66AHH8 pKa = 5.54LRR10 pKa = 11.84YY11 pKa = 10.48SIDD14 pKa = 3.82HH15 pKa = 6.5EE16 pKa = 4.19WLEE19 pKa = 4.7DD20 pKa = 3.84GEE22 pKa = 4.41PACVGITTVAADD34 pKa = 3.59ALGEE38 pKa = 4.37VVFVDD43 pKa = 4.61LPEE46 pKa = 4.76VGAQVEE52 pKa = 4.19AGEE55 pKa = 4.32PCGEE59 pKa = 4.13LEE61 pKa = 4.31STKK64 pKa = 10.66SVSDD68 pKa = 3.46LVSPVSGTVVAVNDD82 pKa = 3.99AVVDD86 pKa = 4.12EE87 pKa = 5.22PGTINEE93 pKa = 4.54DD94 pKa = 3.58PYY96 pKa = 10.65GAGWLFTVTVSAEE109 pKa = 3.89GDD111 pKa = 3.57LLSAQDD117 pKa = 3.83YY118 pKa = 10.85ADD120 pKa = 4.14KK121 pKa = 10.84FDD123 pKa = 4.24AVVDD127 pKa = 4.13GG128 pKa = 4.64
MM1 pKa = 7.03AQSVHH6 pKa = 4.66AHH8 pKa = 5.54LRR10 pKa = 11.84YY11 pKa = 10.48SIDD14 pKa = 3.82HH15 pKa = 6.5EE16 pKa = 4.19WLEE19 pKa = 4.7DD20 pKa = 3.84GEE22 pKa = 4.41PACVGITTVAADD34 pKa = 3.59ALGEE38 pKa = 4.37VVFVDD43 pKa = 4.61LPEE46 pKa = 4.76VGAQVEE52 pKa = 4.19AGEE55 pKa = 4.32PCGEE59 pKa = 4.13LEE61 pKa = 4.31STKK64 pKa = 10.66SVSDD68 pKa = 3.46LVSPVSGTVVAVNDD82 pKa = 3.99AVVDD86 pKa = 4.12EE87 pKa = 5.22PGTINEE93 pKa = 4.54DD94 pKa = 3.58PYY96 pKa = 10.65GAGWLFTVTVSAEE109 pKa = 3.89GDD111 pKa = 3.57LLSAQDD117 pKa = 3.83YY118 pKa = 10.85ADD120 pKa = 4.14KK121 pKa = 10.84FDD123 pKa = 4.24AVVDD127 pKa = 4.13GG128 pKa = 4.64
Molecular weight: 13.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F9PGZ8|F9PGZ8_9ACTO FeS assembly protein SufD OS=Actinomyces sp. oral taxon 175 str. F0384 OX=944560 GN=sufD PE=3 SV=1
MM1 pKa = 8.02PEE3 pKa = 3.85TTFACPDD10 pKa = 3.17LTTFLGLDD18 pKa = 3.6ALGLTAVGQLLTSTRR33 pKa = 11.84AIVEE37 pKa = 3.89CRR39 pKa = 11.84MPIGFEE45 pKa = 4.53DD46 pKa = 4.54PFCRR50 pKa = 11.84ACGAQGVSRR59 pKa = 11.84GTVARR64 pKa = 11.84RR65 pKa = 11.84LAHH68 pKa = 6.45VPVGWRR74 pKa = 11.84PTQLVVRR81 pKa = 11.84VRR83 pKa = 11.84RR84 pKa = 11.84FACTHH89 pKa = 5.79CRR91 pKa = 11.84RR92 pKa = 11.84VWRR95 pKa = 11.84QDD97 pKa = 2.98TSGLAEE103 pKa = 3.78PRR105 pKa = 11.84ARR107 pKa = 11.84LTRR110 pKa = 11.84SAVEE114 pKa = 3.43WGLRR118 pKa = 11.84SLALEE123 pKa = 4.3CMSVSRR129 pKa = 11.84VAAALGISWHH139 pKa = 5.43TANNAILTRR148 pKa = 11.84AEE150 pKa = 4.13QILNEE155 pKa = 4.21APDD158 pKa = 3.4RR159 pKa = 11.84FKK161 pKa = 11.13GVEE164 pKa = 3.7ALGVDD169 pKa = 3.49EE170 pKa = 5.09HH171 pKa = 6.13VWRR174 pKa = 11.84HH175 pKa = 4.17TRR177 pKa = 11.84RR178 pKa = 11.84GDD180 pKa = 3.35RR181 pKa = 11.84YY182 pKa = 7.78VTVMIDD188 pKa = 3.35LTPVRR193 pKa = 11.84DD194 pKa = 3.45RR195 pKa = 11.84SGPARR200 pKa = 11.84LLDD203 pKa = 3.8VVPGRR208 pKa = 11.84SKK210 pKa = 10.88KK211 pKa = 9.31VLKK214 pKa = 9.35TWLSQRR220 pKa = 11.84DD221 pKa = 3.69QEE223 pKa = 3.99WRR225 pKa = 11.84GRR227 pKa = 11.84VEE229 pKa = 4.15VVAMDD234 pKa = 4.21GFTGFKK240 pKa = 10.09SAAGEE245 pKa = 4.05EE246 pKa = 4.49LPQARR251 pKa = 11.84AVMDD255 pKa = 4.41PFHH258 pKa = 7.11VVALAARR265 pKa = 11.84KK266 pKa = 9.32LDD268 pKa = 3.27QCRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84IQRR276 pKa = 11.84AITGRR281 pKa = 11.84RR282 pKa = 11.84GRR284 pKa = 11.84AGDD287 pKa = 3.26RR288 pKa = 11.84LYY290 pKa = 10.75RR291 pKa = 11.84ARR293 pKa = 11.84RR294 pKa = 11.84TLLTGAGLLTDD305 pKa = 4.28AQAEE309 pKa = 4.08RR310 pKa = 11.84LEE312 pKa = 4.11NLFADD317 pKa = 4.11EE318 pKa = 3.8RR319 pKa = 11.84HH320 pKa = 6.13AAVQAAWGVYY330 pKa = 7.36QRR332 pKa = 11.84LIQAYY337 pKa = 7.74RR338 pKa = 11.84TEE340 pKa = 4.01EE341 pKa = 4.11AGLGKK346 pKa = 10.67YY347 pKa = 10.32LMQRR351 pKa = 11.84LIDD354 pKa = 3.9SLRR357 pKa = 11.84QAVPDD362 pKa = 3.89GLEE365 pKa = 4.33EE366 pKa = 4.08IQTLARR372 pKa = 11.84TLTEE376 pKa = 3.82RR377 pKa = 11.84ASDD380 pKa = 2.91ILAYY384 pKa = 9.92FDD386 pKa = 5.12RR387 pKa = 11.84PRR389 pKa = 11.84TSNGPTEE396 pKa = 4.92AINGRR401 pKa = 11.84LEE403 pKa = 3.89QLRR406 pKa = 11.84GIALGFRR413 pKa = 11.84NLTHH417 pKa = 4.71YY418 pKa = 8.32TIRR421 pKa = 11.84SLIHH425 pKa = 5.41TGRR428 pKa = 11.84LKK430 pKa = 11.03DD431 pKa = 3.65HH432 pKa = 6.99LAATTT437 pKa = 3.53
MM1 pKa = 8.02PEE3 pKa = 3.85TTFACPDD10 pKa = 3.17LTTFLGLDD18 pKa = 3.6ALGLTAVGQLLTSTRR33 pKa = 11.84AIVEE37 pKa = 3.89CRR39 pKa = 11.84MPIGFEE45 pKa = 4.53DD46 pKa = 4.54PFCRR50 pKa = 11.84ACGAQGVSRR59 pKa = 11.84GTVARR64 pKa = 11.84RR65 pKa = 11.84LAHH68 pKa = 6.45VPVGWRR74 pKa = 11.84PTQLVVRR81 pKa = 11.84VRR83 pKa = 11.84RR84 pKa = 11.84FACTHH89 pKa = 5.79CRR91 pKa = 11.84RR92 pKa = 11.84VWRR95 pKa = 11.84QDD97 pKa = 2.98TSGLAEE103 pKa = 3.78PRR105 pKa = 11.84ARR107 pKa = 11.84LTRR110 pKa = 11.84SAVEE114 pKa = 3.43WGLRR118 pKa = 11.84SLALEE123 pKa = 4.3CMSVSRR129 pKa = 11.84VAAALGISWHH139 pKa = 5.43TANNAILTRR148 pKa = 11.84AEE150 pKa = 4.13QILNEE155 pKa = 4.21APDD158 pKa = 3.4RR159 pKa = 11.84FKK161 pKa = 11.13GVEE164 pKa = 3.7ALGVDD169 pKa = 3.49EE170 pKa = 5.09HH171 pKa = 6.13VWRR174 pKa = 11.84HH175 pKa = 4.17TRR177 pKa = 11.84RR178 pKa = 11.84GDD180 pKa = 3.35RR181 pKa = 11.84YY182 pKa = 7.78VTVMIDD188 pKa = 3.35LTPVRR193 pKa = 11.84DD194 pKa = 3.45RR195 pKa = 11.84SGPARR200 pKa = 11.84LLDD203 pKa = 3.8VVPGRR208 pKa = 11.84SKK210 pKa = 10.88KK211 pKa = 9.31VLKK214 pKa = 9.35TWLSQRR220 pKa = 11.84DD221 pKa = 3.69QEE223 pKa = 3.99WRR225 pKa = 11.84GRR227 pKa = 11.84VEE229 pKa = 4.15VVAMDD234 pKa = 4.21GFTGFKK240 pKa = 10.09SAAGEE245 pKa = 4.05EE246 pKa = 4.49LPQARR251 pKa = 11.84AVMDD255 pKa = 4.41PFHH258 pKa = 7.11VVALAARR265 pKa = 11.84KK266 pKa = 9.32LDD268 pKa = 3.27QCRR271 pKa = 11.84RR272 pKa = 11.84RR273 pKa = 11.84IQRR276 pKa = 11.84AITGRR281 pKa = 11.84RR282 pKa = 11.84GRR284 pKa = 11.84AGDD287 pKa = 3.26RR288 pKa = 11.84LYY290 pKa = 10.75RR291 pKa = 11.84ARR293 pKa = 11.84RR294 pKa = 11.84TLLTGAGLLTDD305 pKa = 4.28AQAEE309 pKa = 4.08RR310 pKa = 11.84LEE312 pKa = 4.11NLFADD317 pKa = 4.11EE318 pKa = 3.8RR319 pKa = 11.84HH320 pKa = 6.13AAVQAAWGVYY330 pKa = 7.36QRR332 pKa = 11.84LIQAYY337 pKa = 7.74RR338 pKa = 11.84TEE340 pKa = 4.01EE341 pKa = 4.11AGLGKK346 pKa = 10.67YY347 pKa = 10.32LMQRR351 pKa = 11.84LIDD354 pKa = 3.9SLRR357 pKa = 11.84QAVPDD362 pKa = 3.89GLEE365 pKa = 4.33EE366 pKa = 4.08IQTLARR372 pKa = 11.84TLTEE376 pKa = 3.82RR377 pKa = 11.84ASDD380 pKa = 2.91ILAYY384 pKa = 9.92FDD386 pKa = 5.12RR387 pKa = 11.84PRR389 pKa = 11.84TSNGPTEE396 pKa = 4.92AINGRR401 pKa = 11.84LEE403 pKa = 3.89QLRR406 pKa = 11.84GIALGFRR413 pKa = 11.84NLTHH417 pKa = 4.71YY418 pKa = 8.32TIRR421 pKa = 11.84SLIHH425 pKa = 5.41TGRR428 pKa = 11.84LKK430 pKa = 11.03DD431 pKa = 3.65HH432 pKa = 6.99LAATTT437 pKa = 3.53
Molecular weight: 48.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
910945 |
35 |
3150 |
345.3 |
36.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.07 ± 0.072 | 0.776 ± 0.013 |
5.788 ± 0.035 | 5.725 ± 0.044 |
2.485 ± 0.027 | 8.92 ± 0.049 |
2.107 ± 0.022 | 3.963 ± 0.033 |
2.18 ± 0.036 | 10.025 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.007 ± 0.021 | 1.956 ± 0.031 |
5.706 ± 0.04 | 3.181 ± 0.026 |
7.281 ± 0.057 | 6.546 ± 0.04 |
6.399 ± 0.038 | 8.476 ± 0.045 |
1.468 ± 0.024 | 1.94 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
