
Bacteroidales bacterium Barb7
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; unclassified Bacteroidales
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
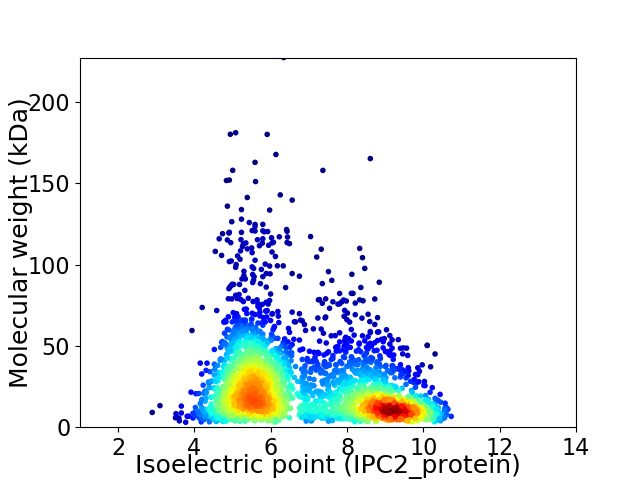
Virtual 2D-PAGE plot for 2857 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
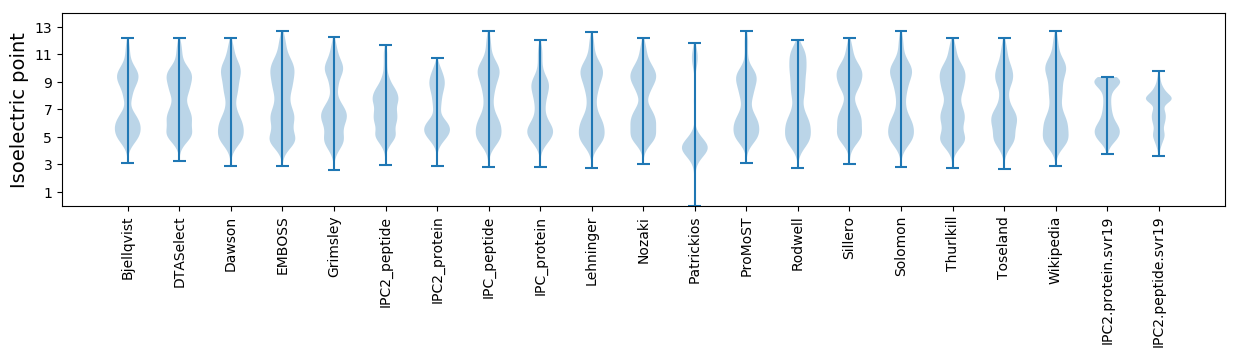
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A180FT70|A0A180FT70_9BACT Xylose isomerase OS=Bacteroidales bacterium Barb7 OX=1633203 GN=xylA PE=3 SV=1
MM1 pKa = 7.36KK2 pKa = 10.63NEE4 pKa = 3.98AQHH7 pKa = 6.82LIDD10 pKa = 4.57RR11 pKa = 11.84FLSARR16 pKa = 11.84NEE18 pKa = 3.66GRR20 pKa = 11.84EE21 pKa = 4.05AYY23 pKa = 10.27FDD25 pKa = 3.95ADD27 pKa = 4.41EE28 pKa = 5.68IDD30 pKa = 5.41DD31 pKa = 4.36LLEE34 pKa = 4.34GCEE37 pKa = 4.16DD38 pKa = 4.24KK39 pKa = 11.37EE40 pKa = 5.27DD41 pKa = 3.82FDD43 pKa = 5.86LYY45 pKa = 11.38EE46 pKa = 4.26EE47 pKa = 4.32VLALGLKK54 pKa = 9.13LHH56 pKa = 7.11PEE58 pKa = 4.33NIGLQTKK65 pKa = 9.86RR66 pKa = 11.84CWQYY70 pKa = 11.12VVEE73 pKa = 4.48EE74 pKa = 4.47NYY76 pKa = 7.72TTALEE81 pKa = 4.59LIEE84 pKa = 4.7QIGATGDD91 pKa = 3.54QEE93 pKa = 4.21IEE95 pKa = 4.05AVQLEE100 pKa = 5.14CYY102 pKa = 10.62LMLDD106 pKa = 3.52MTDD109 pKa = 3.25EE110 pKa = 4.56AEE112 pKa = 4.27ALSEE116 pKa = 4.18KK117 pKa = 9.97MIAEE121 pKa = 4.3GRR123 pKa = 11.84EE124 pKa = 4.03SMDD127 pKa = 3.76LVFEE131 pKa = 4.61YY132 pKa = 10.18IVPLLNDD139 pKa = 3.11KK140 pKa = 10.7EE141 pKa = 4.44MYY143 pKa = 10.43SLALEE148 pKa = 4.23YY149 pKa = 11.05VNRR152 pKa = 11.84GLEE155 pKa = 4.33LYY157 pKa = 9.93PDD159 pKa = 3.69SLVLKK164 pKa = 10.56EE165 pKa = 4.13EE166 pKa = 4.38LCFLMEE172 pKa = 3.99VAEE175 pKa = 4.55DD176 pKa = 3.79LEE178 pKa = 4.72GAIDD182 pKa = 4.26LCNAMIDD189 pKa = 3.83EE190 pKa = 5.04DD191 pKa = 4.66PYY193 pKa = 11.43SYY195 pKa = 10.78EE196 pKa = 3.66YY197 pKa = 10.55WFTLGRR203 pKa = 11.84LYY205 pKa = 11.1SLTEE209 pKa = 4.01EE210 pKa = 4.18FEE212 pKa = 4.21KK213 pKa = 11.1AIDD216 pKa = 3.9AFDD219 pKa = 4.04FAQTCEE225 pKa = 4.32EE226 pKa = 4.56PDD228 pKa = 4.07SEE230 pKa = 5.08LKK232 pKa = 10.63LLKK235 pKa = 10.16AYY237 pKa = 10.56CFFMSDD243 pKa = 3.44KK244 pKa = 11.2YY245 pKa = 11.26EE246 pKa = 4.19EE247 pKa = 4.11ALKK250 pKa = 10.78LYY252 pKa = 10.34TEE254 pKa = 4.28LLKK257 pKa = 11.11DD258 pKa = 3.57EE259 pKa = 4.9EE260 pKa = 4.76KK261 pKa = 11.02NGEE264 pKa = 4.23DD265 pKa = 5.15GEE267 pKa = 5.07GDD269 pKa = 3.71DD270 pKa = 5.24YY271 pKa = 11.39ILEE274 pKa = 4.22RR275 pKa = 11.84VRR277 pKa = 11.84PLLAEE282 pKa = 4.56CYY284 pKa = 8.12MKK286 pKa = 11.06LFDD289 pKa = 5.07YY290 pKa = 10.03EE291 pKa = 3.82AAYY294 pKa = 10.48LILHH298 pKa = 6.42EE299 pKa = 5.64LIADD303 pKa = 3.87DD304 pKa = 5.48AEE306 pKa = 4.31AAGYY310 pKa = 7.57EE311 pKa = 4.3TDD313 pKa = 4.13NADD316 pKa = 3.77TEE318 pKa = 4.86VNGADD323 pKa = 3.87TEE325 pKa = 4.76ADD327 pKa = 3.69DD328 pKa = 4.69TDD330 pKa = 3.98TDD332 pKa = 3.76STDD335 pKa = 3.33AARR338 pKa = 11.84KK339 pKa = 9.87DD340 pKa = 3.69YY341 pKa = 11.22SPYY344 pKa = 9.79IYY346 pKa = 10.9YY347 pKa = 7.49MHH349 pKa = 7.46CCAQTDD355 pKa = 3.95RR356 pKa = 11.84KK357 pKa = 10.22QDD359 pKa = 3.38ALNALEE365 pKa = 4.11TAATVFDD372 pKa = 4.58GSAHH376 pKa = 5.66VLSLMAYY383 pKa = 9.82AYY385 pKa = 9.66MEE387 pKa = 4.27SAEE390 pKa = 4.29NDD392 pKa = 3.58DD393 pKa = 4.75NLDD396 pKa = 3.78VINKK400 pKa = 9.28LFEE403 pKa = 5.87LIDD406 pKa = 4.32DD407 pKa = 5.01PGDD410 pKa = 4.04DD411 pKa = 4.94LEE413 pKa = 5.26EE414 pKa = 4.38EE415 pKa = 4.41ADD417 pKa = 3.71GDD419 pKa = 4.07NPSGKK424 pKa = 9.96DD425 pKa = 3.45GFSEE429 pKa = 4.73TIGKK433 pKa = 8.06MLEE436 pKa = 4.16YY437 pKa = 10.24FKK439 pKa = 10.7KK440 pKa = 10.29IYY442 pKa = 9.9AANPDD447 pKa = 3.25QPYY450 pKa = 10.56INLYY454 pKa = 10.09LATAYY459 pKa = 10.62LINDD463 pKa = 3.48DD464 pKa = 3.65MQNFEE469 pKa = 3.9KK470 pKa = 10.44HH471 pKa = 5.65YY472 pKa = 9.78NLLSAEE478 pKa = 4.34EE479 pKa = 4.08LDD481 pKa = 5.01SYY483 pKa = 10.26AKK485 pKa = 10.26KK486 pKa = 10.91YY487 pKa = 9.94NAGDD491 pKa = 3.44SRR493 pKa = 11.84ALRR496 pKa = 11.84TIAEE500 pKa = 4.12KK501 pKa = 10.66HH502 pKa = 5.34IGLRR506 pKa = 11.84DD507 pKa = 3.35LAEE510 pKa = 4.63EE511 pKa = 4.16YY512 pKa = 10.61LSKK515 pKa = 10.93PEE517 pKa = 3.94NSNN520 pKa = 3.08
MM1 pKa = 7.36KK2 pKa = 10.63NEE4 pKa = 3.98AQHH7 pKa = 6.82LIDD10 pKa = 4.57RR11 pKa = 11.84FLSARR16 pKa = 11.84NEE18 pKa = 3.66GRR20 pKa = 11.84EE21 pKa = 4.05AYY23 pKa = 10.27FDD25 pKa = 3.95ADD27 pKa = 4.41EE28 pKa = 5.68IDD30 pKa = 5.41DD31 pKa = 4.36LLEE34 pKa = 4.34GCEE37 pKa = 4.16DD38 pKa = 4.24KK39 pKa = 11.37EE40 pKa = 5.27DD41 pKa = 3.82FDD43 pKa = 5.86LYY45 pKa = 11.38EE46 pKa = 4.26EE47 pKa = 4.32VLALGLKK54 pKa = 9.13LHH56 pKa = 7.11PEE58 pKa = 4.33NIGLQTKK65 pKa = 9.86RR66 pKa = 11.84CWQYY70 pKa = 11.12VVEE73 pKa = 4.48EE74 pKa = 4.47NYY76 pKa = 7.72TTALEE81 pKa = 4.59LIEE84 pKa = 4.7QIGATGDD91 pKa = 3.54QEE93 pKa = 4.21IEE95 pKa = 4.05AVQLEE100 pKa = 5.14CYY102 pKa = 10.62LMLDD106 pKa = 3.52MTDD109 pKa = 3.25EE110 pKa = 4.56AEE112 pKa = 4.27ALSEE116 pKa = 4.18KK117 pKa = 9.97MIAEE121 pKa = 4.3GRR123 pKa = 11.84EE124 pKa = 4.03SMDD127 pKa = 3.76LVFEE131 pKa = 4.61YY132 pKa = 10.18IVPLLNDD139 pKa = 3.11KK140 pKa = 10.7EE141 pKa = 4.44MYY143 pKa = 10.43SLALEE148 pKa = 4.23YY149 pKa = 11.05VNRR152 pKa = 11.84GLEE155 pKa = 4.33LYY157 pKa = 9.93PDD159 pKa = 3.69SLVLKK164 pKa = 10.56EE165 pKa = 4.13EE166 pKa = 4.38LCFLMEE172 pKa = 3.99VAEE175 pKa = 4.55DD176 pKa = 3.79LEE178 pKa = 4.72GAIDD182 pKa = 4.26LCNAMIDD189 pKa = 3.83EE190 pKa = 5.04DD191 pKa = 4.66PYY193 pKa = 11.43SYY195 pKa = 10.78EE196 pKa = 3.66YY197 pKa = 10.55WFTLGRR203 pKa = 11.84LYY205 pKa = 11.1SLTEE209 pKa = 4.01EE210 pKa = 4.18FEE212 pKa = 4.21KK213 pKa = 11.1AIDD216 pKa = 3.9AFDD219 pKa = 4.04FAQTCEE225 pKa = 4.32EE226 pKa = 4.56PDD228 pKa = 4.07SEE230 pKa = 5.08LKK232 pKa = 10.63LLKK235 pKa = 10.16AYY237 pKa = 10.56CFFMSDD243 pKa = 3.44KK244 pKa = 11.2YY245 pKa = 11.26EE246 pKa = 4.19EE247 pKa = 4.11ALKK250 pKa = 10.78LYY252 pKa = 10.34TEE254 pKa = 4.28LLKK257 pKa = 11.11DD258 pKa = 3.57EE259 pKa = 4.9EE260 pKa = 4.76KK261 pKa = 11.02NGEE264 pKa = 4.23DD265 pKa = 5.15GEE267 pKa = 5.07GDD269 pKa = 3.71DD270 pKa = 5.24YY271 pKa = 11.39ILEE274 pKa = 4.22RR275 pKa = 11.84VRR277 pKa = 11.84PLLAEE282 pKa = 4.56CYY284 pKa = 8.12MKK286 pKa = 11.06LFDD289 pKa = 5.07YY290 pKa = 10.03EE291 pKa = 3.82AAYY294 pKa = 10.48LILHH298 pKa = 6.42EE299 pKa = 5.64LIADD303 pKa = 3.87DD304 pKa = 5.48AEE306 pKa = 4.31AAGYY310 pKa = 7.57EE311 pKa = 4.3TDD313 pKa = 4.13NADD316 pKa = 3.77TEE318 pKa = 4.86VNGADD323 pKa = 3.87TEE325 pKa = 4.76ADD327 pKa = 3.69DD328 pKa = 4.69TDD330 pKa = 3.98TDD332 pKa = 3.76STDD335 pKa = 3.33AARR338 pKa = 11.84KK339 pKa = 9.87DD340 pKa = 3.69YY341 pKa = 11.22SPYY344 pKa = 9.79IYY346 pKa = 10.9YY347 pKa = 7.49MHH349 pKa = 7.46CCAQTDD355 pKa = 3.95RR356 pKa = 11.84KK357 pKa = 10.22QDD359 pKa = 3.38ALNALEE365 pKa = 4.11TAATVFDD372 pKa = 4.58GSAHH376 pKa = 5.66VLSLMAYY383 pKa = 9.82AYY385 pKa = 9.66MEE387 pKa = 4.27SAEE390 pKa = 4.29NDD392 pKa = 3.58DD393 pKa = 4.75NLDD396 pKa = 3.78VINKK400 pKa = 9.28LFEE403 pKa = 5.87LIDD406 pKa = 4.32DD407 pKa = 5.01PGDD410 pKa = 4.04DD411 pKa = 4.94LEE413 pKa = 5.26EE414 pKa = 4.38EE415 pKa = 4.41ADD417 pKa = 3.71GDD419 pKa = 4.07NPSGKK424 pKa = 9.96DD425 pKa = 3.45GFSEE429 pKa = 4.73TIGKK433 pKa = 8.06MLEE436 pKa = 4.16YY437 pKa = 10.24FKK439 pKa = 10.7KK440 pKa = 10.29IYY442 pKa = 9.9AANPDD447 pKa = 3.25QPYY450 pKa = 10.56INLYY454 pKa = 10.09LATAYY459 pKa = 10.62LINDD463 pKa = 3.48DD464 pKa = 3.65MQNFEE469 pKa = 3.9KK470 pKa = 10.44HH471 pKa = 5.65YY472 pKa = 9.78NLLSAEE478 pKa = 4.34EE479 pKa = 4.08LDD481 pKa = 5.01SYY483 pKa = 10.26AKK485 pKa = 10.26KK486 pKa = 10.91YY487 pKa = 9.94NAGDD491 pKa = 3.44SRR493 pKa = 11.84ALRR496 pKa = 11.84TIAEE500 pKa = 4.12KK501 pKa = 10.66HH502 pKa = 5.34IGLRR506 pKa = 11.84DD507 pKa = 3.35LAEE510 pKa = 4.63EE511 pKa = 4.16YY512 pKa = 10.61LSKK515 pKa = 10.93PEE517 pKa = 3.94NSNN520 pKa = 3.08
Molecular weight: 59.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A180FTK7|A0A180FTK7_9BACT Capsular glucan synthase OS=Bacteroidales bacterium Barb7 OX=1633203 GN=glgA PE=4 SV=1
MM1 pKa = 7.26LTGDD5 pKa = 4.07APPYY9 pKa = 10.4GGVRR13 pKa = 11.84LCRR16 pKa = 11.84VAVYY20 pKa = 8.22GQPVLKK26 pKa = 10.36EE27 pKa = 3.82LLLVSEE33 pKa = 4.68NILRR37 pKa = 11.84YY38 pKa = 8.42FTEE41 pKa = 3.77IDD43 pKa = 3.38IEE45 pKa = 4.42VAPLRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84IINEE56 pKa = 3.89RR57 pKa = 11.84VKK59 pKa = 10.8HH60 pKa = 5.67PKK62 pKa = 10.15LDD64 pKa = 3.34ILNVCRR70 pKa = 11.84FKK72 pKa = 11.24VALLDD77 pKa = 4.49FPHH80 pKa = 6.47HH81 pKa = 6.35TPPAFLRR88 pKa = 11.84LGKK91 pKa = 8.93RR92 pKa = 11.84TVGIDD97 pKa = 3.27VVGIEE102 pKa = 4.59VIGPALIRR110 pKa = 11.84IIRR113 pKa = 11.84HH114 pKa = 3.76IQYY117 pKa = 8.65PQRR120 pKa = 11.84GSIAYY125 pKa = 9.43DD126 pKa = 3.33LLLVRR131 pKa = 11.84EE132 pKa = 4.45KK133 pKa = 10.84LLHH136 pKa = 6.62FDD138 pKa = 3.17GTHH141 pKa = 4.47IVVRR145 pKa = 11.84LVVFGDD151 pKa = 3.46MAGNGVGVFARR162 pKa = 11.84KK163 pKa = 8.04YY164 pKa = 7.87TVNAVPSRR172 pKa = 11.84QGAVLAVVNVVGKK185 pKa = 10.52SGFGEE190 pKa = 4.05AGRR193 pKa = 11.84GKK195 pKa = 9.94VVVRR199 pKa = 11.84RR200 pKa = 11.84GRR202 pKa = 11.84DD203 pKa = 3.53LPCVGVLQAGVDD215 pKa = 3.68LRR217 pKa = 11.84AFKK220 pKa = 10.23IIKK223 pKa = 10.17VGILNVAACVLALVMTCNQIGKK245 pKa = 9.12LRR247 pKa = 11.84LKK249 pKa = 10.63GYY251 pKa = 9.35LARR254 pKa = 11.84TGQRR258 pKa = 11.84QQRR261 pKa = 11.84QLVEE265 pKa = 3.77QVGQLLALRR274 pKa = 11.84LVGCVQPPQQIINRR288 pKa = 11.84LIPYY292 pKa = 9.75GSLHH296 pKa = 5.82RR297 pKa = 11.84HH298 pKa = 4.56RR299 pKa = 11.84HH300 pKa = 4.61LQHH303 pKa = 5.3VHH305 pKa = 6.18HH306 pKa = 6.86GAAQRR311 pKa = 11.84EE312 pKa = 4.21ILIEE316 pKa = 4.24LIIHH320 pKa = 5.83IHH322 pKa = 5.61AKK324 pKa = 9.9HH325 pKa = 6.1RR326 pKa = 11.84LALHH330 pKa = 5.92TEE332 pKa = 3.95HH333 pKa = 6.98GLVFQRR339 pKa = 11.84NTDD342 pKa = 3.24RR343 pKa = 11.84RR344 pKa = 11.84IACHH348 pKa = 5.77NALVEE353 pKa = 4.36DD354 pKa = 4.74CNRR357 pKa = 11.84PHH359 pKa = 7.62DD360 pKa = 4.57IIHH363 pKa = 6.74RR364 pKa = 11.84IIRR367 pKa = 11.84ILGKK371 pKa = 10.07LCPAGCHH378 pKa = 6.24PYY380 pKa = 10.42RR381 pKa = 11.84PASHH385 pKa = 6.68IRR387 pKa = 11.84CPQLNLRR394 pKa = 11.84AA395 pKa = 4.03
MM1 pKa = 7.26LTGDD5 pKa = 4.07APPYY9 pKa = 10.4GGVRR13 pKa = 11.84LCRR16 pKa = 11.84VAVYY20 pKa = 8.22GQPVLKK26 pKa = 10.36EE27 pKa = 3.82LLLVSEE33 pKa = 4.68NILRR37 pKa = 11.84YY38 pKa = 8.42FTEE41 pKa = 3.77IDD43 pKa = 3.38IEE45 pKa = 4.42VAPLRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84IINEE56 pKa = 3.89RR57 pKa = 11.84VKK59 pKa = 10.8HH60 pKa = 5.67PKK62 pKa = 10.15LDD64 pKa = 3.34ILNVCRR70 pKa = 11.84FKK72 pKa = 11.24VALLDD77 pKa = 4.49FPHH80 pKa = 6.47HH81 pKa = 6.35TPPAFLRR88 pKa = 11.84LGKK91 pKa = 8.93RR92 pKa = 11.84TVGIDD97 pKa = 3.27VVGIEE102 pKa = 4.59VIGPALIRR110 pKa = 11.84IIRR113 pKa = 11.84HH114 pKa = 3.76IQYY117 pKa = 8.65PQRR120 pKa = 11.84GSIAYY125 pKa = 9.43DD126 pKa = 3.33LLLVRR131 pKa = 11.84EE132 pKa = 4.45KK133 pKa = 10.84LLHH136 pKa = 6.62FDD138 pKa = 3.17GTHH141 pKa = 4.47IVVRR145 pKa = 11.84LVVFGDD151 pKa = 3.46MAGNGVGVFARR162 pKa = 11.84KK163 pKa = 8.04YY164 pKa = 7.87TVNAVPSRR172 pKa = 11.84QGAVLAVVNVVGKK185 pKa = 10.52SGFGEE190 pKa = 4.05AGRR193 pKa = 11.84GKK195 pKa = 9.94VVVRR199 pKa = 11.84RR200 pKa = 11.84GRR202 pKa = 11.84DD203 pKa = 3.53LPCVGVLQAGVDD215 pKa = 3.68LRR217 pKa = 11.84AFKK220 pKa = 10.23IIKK223 pKa = 10.17VGILNVAACVLALVMTCNQIGKK245 pKa = 9.12LRR247 pKa = 11.84LKK249 pKa = 10.63GYY251 pKa = 9.35LARR254 pKa = 11.84TGQRR258 pKa = 11.84QQRR261 pKa = 11.84QLVEE265 pKa = 3.77QVGQLLALRR274 pKa = 11.84LVGCVQPPQQIINRR288 pKa = 11.84LIPYY292 pKa = 9.75GSLHH296 pKa = 5.82RR297 pKa = 11.84HH298 pKa = 4.56RR299 pKa = 11.84HH300 pKa = 4.61LQHH303 pKa = 5.3VHH305 pKa = 6.18HH306 pKa = 6.86GAAQRR311 pKa = 11.84EE312 pKa = 4.21ILIEE316 pKa = 4.24LIIHH320 pKa = 5.83IHH322 pKa = 5.61AKK324 pKa = 9.9HH325 pKa = 6.1RR326 pKa = 11.84LALHH330 pKa = 5.92TEE332 pKa = 3.95HH333 pKa = 6.98GLVFQRR339 pKa = 11.84NTDD342 pKa = 3.24RR343 pKa = 11.84RR344 pKa = 11.84IACHH348 pKa = 5.77NALVEE353 pKa = 4.36DD354 pKa = 4.74CNRR357 pKa = 11.84PHH359 pKa = 7.62DD360 pKa = 4.57IIHH363 pKa = 6.74RR364 pKa = 11.84IIRR367 pKa = 11.84ILGKK371 pKa = 10.07LCPAGCHH378 pKa = 6.24PYY380 pKa = 10.42RR381 pKa = 11.84PASHH385 pKa = 6.68IRR387 pKa = 11.84CPQLNLRR394 pKa = 11.84AA395 pKa = 4.03
Molecular weight: 44.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
695753 |
29 |
2053 |
243.5 |
27.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.637 ± 0.066 | 1.324 ± 0.025 |
5.301 ± 0.036 | 6.184 ± 0.05 |
4.943 ± 0.039 | 7.098 ± 0.06 |
2.067 ± 0.025 | 6.875 ± 0.057 |
6.211 ± 0.057 | 9.225 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.29 ± 0.027 | 4.807 ± 0.047 |
3.844 ± 0.036 | 3.323 ± 0.028 |
5.131 ± 0.043 | 6.323 ± 0.047 |
5.645 ± 0.043 | 6.774 ± 0.056 |
0.977 ± 0.019 | 4.021 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
