
Seal anellovirus TFFN/USA/2006
Taxonomy: Viruses; Anelloviridae; unclassified Anelloviridae
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
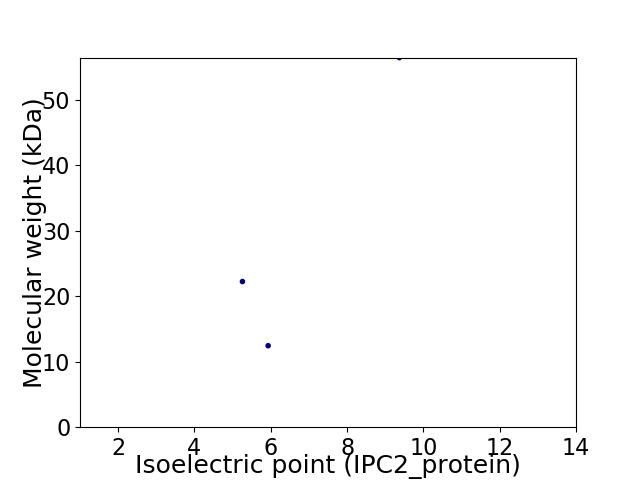
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|F1CHU6|F1CHU6_9VIRU Orf3 OS=Seal anellovirus TFFN/USA/2006 OX=991022 PE=4 SV=1
MM1 pKa = 6.51TTLMTQKK8 pKa = 10.5KK9 pKa = 9.63PMMPEE14 pKa = 3.67QPGLEE19 pKa = 4.52GINIKK24 pKa = 10.11CLHH27 pKa = 6.86ANPILQKK34 pKa = 9.17QAKK37 pKa = 6.81TVIVLTGALSYY48 pKa = 11.45LNAFPCGILGADD60 pKa = 3.66PYY62 pKa = 11.31GLNINSFLSSLEE74 pKa = 3.99TLSTEE79 pKa = 4.07EE80 pKa = 4.47QSAKK84 pKa = 9.34TPKK87 pKa = 9.86TSSLKK92 pKa = 10.25LRR94 pKa = 11.84RR95 pKa = 11.84ANTLKK100 pKa = 10.52EE101 pKa = 4.11KK102 pKa = 10.86YY103 pKa = 9.74RR104 pKa = 11.84LAPLQGGRR112 pKa = 11.84SMRR115 pKa = 11.84RR116 pKa = 11.84TSSEE120 pKa = 4.35GILTPQGSSRR130 pKa = 11.84RR131 pKa = 11.84EE132 pKa = 4.01LIEE135 pKa = 4.04EE136 pKa = 4.2LLDD139 pKa = 3.79LVEE142 pKa = 6.18KK143 pKa = 10.78SSQPCWCQKK152 pKa = 10.43EE153 pKa = 4.31SDD155 pKa = 4.02SGEE158 pKa = 4.22EE159 pKa = 3.85TSDD162 pKa = 4.4ASDD165 pKa = 3.57SDD167 pKa = 3.41ISYY170 pKa = 10.8LDD172 pKa = 3.21SWSDD176 pKa = 3.42EE177 pKa = 4.15EE178 pKa = 4.65NPGGEE183 pKa = 4.45GAPLVPPQGGAPPRR197 pKa = 11.84YY198 pKa = 8.66PSQFLFF204 pKa = 4.03
MM1 pKa = 6.51TTLMTQKK8 pKa = 10.5KK9 pKa = 9.63PMMPEE14 pKa = 3.67QPGLEE19 pKa = 4.52GINIKK24 pKa = 10.11CLHH27 pKa = 6.86ANPILQKK34 pKa = 9.17QAKK37 pKa = 6.81TVIVLTGALSYY48 pKa = 11.45LNAFPCGILGADD60 pKa = 3.66PYY62 pKa = 11.31GLNINSFLSSLEE74 pKa = 3.99TLSTEE79 pKa = 4.07EE80 pKa = 4.47QSAKK84 pKa = 9.34TPKK87 pKa = 9.86TSSLKK92 pKa = 10.25LRR94 pKa = 11.84RR95 pKa = 11.84ANTLKK100 pKa = 10.52EE101 pKa = 4.11KK102 pKa = 10.86YY103 pKa = 9.74RR104 pKa = 11.84LAPLQGGRR112 pKa = 11.84SMRR115 pKa = 11.84RR116 pKa = 11.84TSSEE120 pKa = 4.35GILTPQGSSRR130 pKa = 11.84RR131 pKa = 11.84EE132 pKa = 4.01LIEE135 pKa = 4.04EE136 pKa = 4.2LLDD139 pKa = 3.79LVEE142 pKa = 6.18KK143 pKa = 10.78SSQPCWCQKK152 pKa = 10.43EE153 pKa = 4.31SDD155 pKa = 4.02SGEE158 pKa = 4.22EE159 pKa = 3.85TSDD162 pKa = 4.4ASDD165 pKa = 3.57SDD167 pKa = 3.41ISYY170 pKa = 10.8LDD172 pKa = 3.21SWSDD176 pKa = 3.42EE177 pKa = 4.15EE178 pKa = 4.65NPGGEE183 pKa = 4.45GAPLVPPQGGAPPRR197 pKa = 11.84YY198 pKa = 8.66PSQFLFF204 pKa = 4.03
Molecular weight: 22.26 kDa
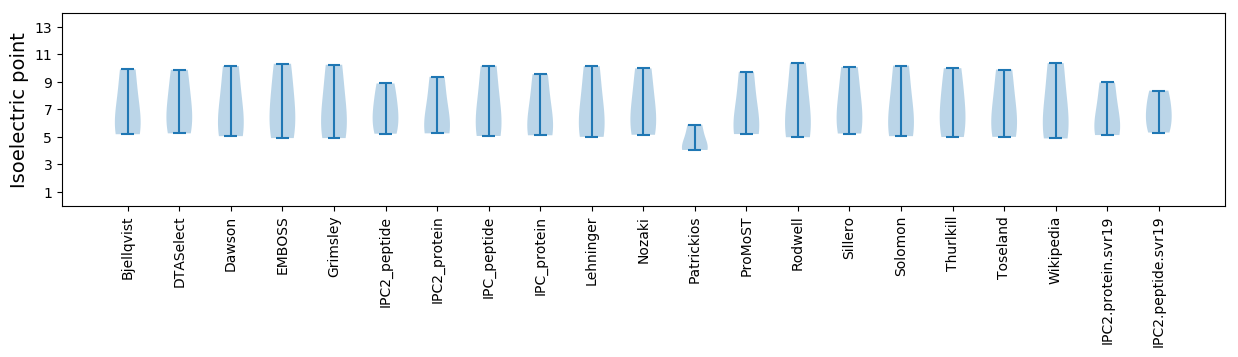
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F1CHU6|F1CHU6_9VIRU Orf3 OS=Seal anellovirus TFFN/USA/2006 OX=991022 PE=4 SV=1
MM1 pKa = 7.47ARR3 pKa = 11.84RR4 pKa = 11.84FRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84FQRR11 pKa = 11.84WRR13 pKa = 11.84RR14 pKa = 11.84GRR16 pKa = 11.84YY17 pKa = 8.06HH18 pKa = 6.89PRR20 pKa = 11.84RR21 pKa = 11.84GYY23 pKa = 9.72RR24 pKa = 11.84RR25 pKa = 11.84WRR27 pKa = 11.84PFHH30 pKa = 6.58HH31 pKa = 5.74WRR33 pKa = 11.84HH34 pKa = 3.62WRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84YY41 pKa = 8.21FRR43 pKa = 11.84HH44 pKa = 4.86RR45 pKa = 11.84TAAVRR50 pKa = 11.84YY51 pKa = 8.65RR52 pKa = 11.84PSRR55 pKa = 11.84RR56 pKa = 11.84HH57 pKa = 5.23KK58 pKa = 10.78VVVCRR63 pKa = 11.84GWEE66 pKa = 3.74PLGNVCKK73 pKa = 9.48EE74 pKa = 4.09TLASQEE80 pKa = 3.79ATPYY84 pKa = 10.88KK85 pKa = 10.43DD86 pKa = 3.68IEE88 pKa = 4.49ADD90 pKa = 3.59YY91 pKa = 11.32DD92 pKa = 3.94PTTSTLDD99 pKa = 3.2QFKK102 pKa = 10.86GSWQGTWGHH111 pKa = 6.63HH112 pKa = 5.01YY113 pKa = 8.0FTLEE117 pKa = 3.81NLVKK121 pKa = 10.67RR122 pKa = 11.84NRR124 pKa = 11.84QLMNTFTSDD133 pKa = 2.47WRR135 pKa = 11.84GWDD138 pKa = 3.39YY139 pKa = 11.44YY140 pKa = 11.31QFLGGVFYY148 pKa = 10.26MPKK151 pKa = 9.68ISGMDD156 pKa = 3.07YY157 pKa = 10.73FIYY160 pKa = 10.54LDD162 pKa = 4.49YY163 pKa = 11.33DD164 pKa = 3.84LQDD167 pKa = 4.03LKK169 pKa = 11.6AKK171 pKa = 10.51GDD173 pKa = 3.51EE174 pKa = 4.1TDD176 pKa = 3.92LYY178 pKa = 11.53KK179 pKa = 10.87HH180 pKa = 5.68MKK182 pKa = 9.33SWIHH186 pKa = 5.89PGIYY190 pKa = 8.58MNRR193 pKa = 11.84YY194 pKa = 6.29GAKK197 pKa = 9.56MITSPHH203 pKa = 6.1RR204 pKa = 11.84TTAPRR209 pKa = 11.84LFRR212 pKa = 11.84KK213 pKa = 9.74VRR215 pKa = 11.84IKK217 pKa = 10.71PPASWEE223 pKa = 3.75GLYY226 pKa = 10.17PINTGFKK233 pKa = 10.82YY234 pKa = 10.31IFCHH238 pKa = 5.69WAWTVFDD245 pKa = 3.62WDD247 pKa = 3.8MPFFDD252 pKa = 4.72SYY254 pKa = 11.8CYY256 pKa = 10.55LKK258 pKa = 10.79SCATKK263 pKa = 10.02PPKK266 pKa = 9.96PMCEE270 pKa = 3.82QCCNEE275 pKa = 3.98RR276 pKa = 11.84PWWSGLKK283 pKa = 10.14DD284 pKa = 3.43SHH286 pKa = 7.01NMCKK290 pKa = 10.25PNFDD294 pKa = 5.7DD295 pKa = 5.06NFNDD299 pKa = 3.85SKK301 pKa = 10.91EE302 pKa = 4.42AYY304 pKa = 8.63DD305 pKa = 5.73ARR307 pKa = 11.84ATWVRR312 pKa = 11.84RR313 pKa = 11.84DD314 pKa = 3.5QYY316 pKa = 11.74QMSSCEE322 pKa = 4.19SNTPEE327 pKa = 4.17TGKK330 pKa = 10.48DD331 pKa = 3.45CNCFNWGPFLPQCFPLWNSWGRR353 pKa = 11.84SVWFKK358 pKa = 11.27YY359 pKa = 10.7KK360 pKa = 10.82LFFKK364 pKa = 10.92FSGDD368 pKa = 3.06SVYY371 pKa = 10.82RR372 pKa = 11.84RR373 pKa = 11.84TVSKK377 pKa = 10.56DD378 pKa = 3.24SKK380 pKa = 11.24DD381 pKa = 4.44LIPEE385 pKa = 4.25APKK388 pKa = 10.74GKK390 pKa = 9.91YY391 pKa = 8.93SQGEE395 pKa = 4.31IPSSSSPRR403 pKa = 11.84RR404 pKa = 11.84PLDD407 pKa = 3.23EE408 pKa = 6.4ADD410 pKa = 3.55ILRR413 pKa = 11.84GDD415 pKa = 4.89LDD417 pKa = 3.68PSGILTEE424 pKa = 3.9RR425 pKa = 11.84AYY427 pKa = 10.8RR428 pKa = 11.84GITRR432 pKa = 11.84PGGEE436 pKa = 4.49EE437 pKa = 4.04QPTMLVPKK445 pKa = 9.89RR446 pKa = 11.84VRR448 pKa = 11.84FRR450 pKa = 11.84RR451 pKa = 11.84GDD453 pKa = 3.08IRR455 pKa = 11.84RR456 pKa = 11.84KK457 pKa = 9.6RR458 pKa = 11.84LRR460 pKa = 11.84HH461 pKa = 4.93LLSRR465 pKa = 11.84LMEE468 pKa = 4.19RR469 pKa = 4.35
MM1 pKa = 7.47ARR3 pKa = 11.84RR4 pKa = 11.84FRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84FQRR11 pKa = 11.84WRR13 pKa = 11.84RR14 pKa = 11.84GRR16 pKa = 11.84YY17 pKa = 8.06HH18 pKa = 6.89PRR20 pKa = 11.84RR21 pKa = 11.84GYY23 pKa = 9.72RR24 pKa = 11.84RR25 pKa = 11.84WRR27 pKa = 11.84PFHH30 pKa = 6.58HH31 pKa = 5.74WRR33 pKa = 11.84HH34 pKa = 3.62WRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84YY41 pKa = 8.21FRR43 pKa = 11.84HH44 pKa = 4.86RR45 pKa = 11.84TAAVRR50 pKa = 11.84YY51 pKa = 8.65RR52 pKa = 11.84PSRR55 pKa = 11.84RR56 pKa = 11.84HH57 pKa = 5.23KK58 pKa = 10.78VVVCRR63 pKa = 11.84GWEE66 pKa = 3.74PLGNVCKK73 pKa = 9.48EE74 pKa = 4.09TLASQEE80 pKa = 3.79ATPYY84 pKa = 10.88KK85 pKa = 10.43DD86 pKa = 3.68IEE88 pKa = 4.49ADD90 pKa = 3.59YY91 pKa = 11.32DD92 pKa = 3.94PTTSTLDD99 pKa = 3.2QFKK102 pKa = 10.86GSWQGTWGHH111 pKa = 6.63HH112 pKa = 5.01YY113 pKa = 8.0FTLEE117 pKa = 3.81NLVKK121 pKa = 10.67RR122 pKa = 11.84NRR124 pKa = 11.84QLMNTFTSDD133 pKa = 2.47WRR135 pKa = 11.84GWDD138 pKa = 3.39YY139 pKa = 11.44YY140 pKa = 11.31QFLGGVFYY148 pKa = 10.26MPKK151 pKa = 9.68ISGMDD156 pKa = 3.07YY157 pKa = 10.73FIYY160 pKa = 10.54LDD162 pKa = 4.49YY163 pKa = 11.33DD164 pKa = 3.84LQDD167 pKa = 4.03LKK169 pKa = 11.6AKK171 pKa = 10.51GDD173 pKa = 3.51EE174 pKa = 4.1TDD176 pKa = 3.92LYY178 pKa = 11.53KK179 pKa = 10.87HH180 pKa = 5.68MKK182 pKa = 9.33SWIHH186 pKa = 5.89PGIYY190 pKa = 8.58MNRR193 pKa = 11.84YY194 pKa = 6.29GAKK197 pKa = 9.56MITSPHH203 pKa = 6.1RR204 pKa = 11.84TTAPRR209 pKa = 11.84LFRR212 pKa = 11.84KK213 pKa = 9.74VRR215 pKa = 11.84IKK217 pKa = 10.71PPASWEE223 pKa = 3.75GLYY226 pKa = 10.17PINTGFKK233 pKa = 10.82YY234 pKa = 10.31IFCHH238 pKa = 5.69WAWTVFDD245 pKa = 3.62WDD247 pKa = 3.8MPFFDD252 pKa = 4.72SYY254 pKa = 11.8CYY256 pKa = 10.55LKK258 pKa = 10.79SCATKK263 pKa = 10.02PPKK266 pKa = 9.96PMCEE270 pKa = 3.82QCCNEE275 pKa = 3.98RR276 pKa = 11.84PWWSGLKK283 pKa = 10.14DD284 pKa = 3.43SHH286 pKa = 7.01NMCKK290 pKa = 10.25PNFDD294 pKa = 5.7DD295 pKa = 5.06NFNDD299 pKa = 3.85SKK301 pKa = 10.91EE302 pKa = 4.42AYY304 pKa = 8.63DD305 pKa = 5.73ARR307 pKa = 11.84ATWVRR312 pKa = 11.84RR313 pKa = 11.84DD314 pKa = 3.5QYY316 pKa = 11.74QMSSCEE322 pKa = 4.19SNTPEE327 pKa = 4.17TGKK330 pKa = 10.48DD331 pKa = 3.45CNCFNWGPFLPQCFPLWNSWGRR353 pKa = 11.84SVWFKK358 pKa = 11.27YY359 pKa = 10.7KK360 pKa = 10.82LFFKK364 pKa = 10.92FSGDD368 pKa = 3.06SVYY371 pKa = 10.82RR372 pKa = 11.84RR373 pKa = 11.84TVSKK377 pKa = 10.56DD378 pKa = 3.24SKK380 pKa = 11.24DD381 pKa = 4.44LIPEE385 pKa = 4.25APKK388 pKa = 10.74GKK390 pKa = 9.91YY391 pKa = 8.93SQGEE395 pKa = 4.31IPSSSSPRR403 pKa = 11.84RR404 pKa = 11.84PLDD407 pKa = 3.23EE408 pKa = 6.4ADD410 pKa = 3.55ILRR413 pKa = 11.84GDD415 pKa = 4.89LDD417 pKa = 3.68PSGILTEE424 pKa = 3.9RR425 pKa = 11.84AYY427 pKa = 10.8RR428 pKa = 11.84GITRR432 pKa = 11.84PGGEE436 pKa = 4.49EE437 pKa = 4.04QPTMLVPKK445 pKa = 9.89RR446 pKa = 11.84VRR448 pKa = 11.84FRR450 pKa = 11.84RR451 pKa = 11.84GDD453 pKa = 3.08IRR455 pKa = 11.84RR456 pKa = 11.84KK457 pKa = 9.6RR458 pKa = 11.84LRR460 pKa = 11.84HH461 pKa = 4.93LLSRR465 pKa = 11.84LMEE468 pKa = 4.19RR469 pKa = 4.35
Molecular weight: 56.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
784 |
111 |
469 |
261.3 |
30.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.592 ± 0.746 | 2.423 ± 0.329 |
5.612 ± 0.749 | 5.23 ± 1.248 |
4.592 ± 1.045 | 7.653 ± 1.594 |
2.296 ± 0.727 | 4.209 ± 1.263 |
5.995 ± 0.49 | 8.291 ± 2.137 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.679 ± 0.094 | 3.444 ± 0.331 |
6.633 ± 1.152 | 3.827 ± 0.98 |
9.056 ± 2.874 | 7.143 ± 2.005 |
5.102 ± 0.729 | 3.444 ± 1.092 |
3.316 ± 1.088 | 4.464 ± 0.999 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
