
Sphingomonas laterariae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
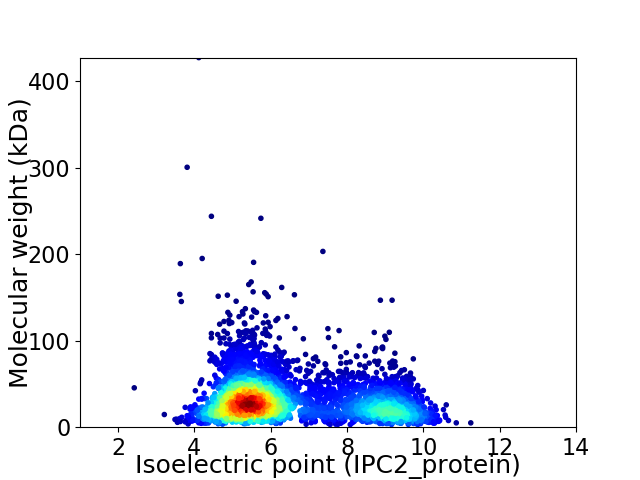
Virtual 2D-PAGE plot for 4276 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A239J1C2|A0A239J1C2_9SPHN Integrase OS=Sphingomonas laterariae OX=861865 GN=SAMN06295912_12918 PE=4 SV=1
MM1 pKa = 7.36TSDD4 pKa = 3.11ILMNAILSLDD14 pKa = 3.68AYY16 pKa = 10.7NRR18 pKa = 11.84GYY20 pKa = 10.87DD21 pKa = 3.2AGVQITEE28 pKa = 4.58PDD30 pKa = 4.52DD31 pKa = 3.16ITPYY35 pKa = 10.71KK36 pKa = 10.33VGNATFLKK44 pKa = 9.49QSSVAEE50 pKa = 4.07SSEE53 pKa = 3.88GVTAGFYY60 pKa = 10.67AIAYY64 pKa = 8.08SYY66 pKa = 11.23NGEE69 pKa = 4.23TVISYY74 pKa = 10.46RR75 pKa = 11.84GTDD78 pKa = 3.81DD79 pKa = 3.57YY80 pKa = 12.03DD81 pKa = 5.27GITSIPSSLDD91 pKa = 3.21AANGWPMGGGSITTAQGRR109 pKa = 11.84MALEE113 pKa = 4.82FYY115 pKa = 10.94KK116 pKa = 10.83DD117 pKa = 3.83VADD120 pKa = 4.48LPYY123 pKa = 10.94GSDD126 pKa = 3.38LRR128 pKa = 11.84NVAITLTGHH137 pKa = 5.85SLGGGLAGYY146 pKa = 8.81VASLYY151 pKa = 9.94GQNADD156 pKa = 4.3IFDD159 pKa = 3.78SMAFKK164 pKa = 10.68LAATATINVAYY175 pKa = 10.02HH176 pKa = 6.1PEE178 pKa = 4.48YY179 pKa = 10.15YY180 pKa = 9.84TDD182 pKa = 3.52PEE184 pKa = 4.74YY185 pKa = 11.09YY186 pKa = 9.7PEE188 pKa = 5.76PIFSDD193 pKa = 3.43AQEE196 pKa = 4.06LVYY199 pKa = 10.84NGYY202 pKa = 7.98STPWEE207 pKa = 4.19NSHH210 pKa = 6.85PEE212 pKa = 3.97YY213 pKa = 11.22VNGYY217 pKa = 8.02EE218 pKa = 5.33LSGQVISEE226 pKa = 4.38EE227 pKa = 3.69NWYY230 pKa = 6.7PTPGVDD236 pKa = 3.19SVGYY240 pKa = 8.25NTYY243 pKa = 11.17DD244 pKa = 5.27DD245 pKa = 5.41GDD247 pKa = 4.03LDD249 pKa = 5.25SNDD252 pKa = 3.46KK253 pKa = 11.03HH254 pKa = 8.04SMSSFVIRR262 pKa = 11.84YY263 pKa = 6.44YY264 pKa = 11.44AEE266 pKa = 3.71VGGANDD272 pKa = 4.01SALSTDD278 pKa = 3.35WQNAAPYY285 pKa = 8.7FWAVLYY291 pKa = 10.49DD292 pKa = 3.84DD293 pKa = 5.3QLAQAIGADD302 pKa = 3.84AKK304 pKa = 11.17DD305 pKa = 3.56GTLKK309 pKa = 11.11DD310 pKa = 3.34EE311 pKa = 4.51GKK313 pKa = 9.86YY314 pKa = 10.81AEE316 pKa = 4.24IVRR319 pKa = 11.84TAIAYY324 pKa = 9.66SSLEE328 pKa = 4.08KK329 pKa = 11.0NVDD332 pKa = 3.14GTGLIFGDD340 pKa = 3.35TGIRR344 pKa = 11.84ALYY347 pKa = 10.61DD348 pKa = 3.52DD349 pKa = 5.58ANDD352 pKa = 3.55LGARR356 pKa = 11.84IADD359 pKa = 4.16PLSGDD364 pKa = 3.86LFDD367 pKa = 4.42TYY369 pKa = 11.01GTVISRR375 pKa = 11.84AFVEE379 pKa = 4.95FAGSLALHH387 pKa = 6.84EE388 pKa = 4.23IEE390 pKa = 4.08KK391 pKa = 10.17TEE393 pKa = 3.83FAGAINGVLSHH404 pKa = 7.57DD405 pKa = 4.14ATNHH409 pKa = 5.21TLTVNYY415 pKa = 10.6ADD417 pKa = 4.87ALWIAAGKK425 pKa = 8.94GANDD429 pKa = 3.76IIVSRR434 pKa = 11.84EE435 pKa = 3.96NIVEE439 pKa = 3.97QAFGVDD445 pKa = 3.49EE446 pKa = 4.48TQLRR450 pKa = 11.84TAMWDD455 pKa = 3.03LWQNDD460 pKa = 3.28GNAVFDD466 pKa = 3.87KK467 pKa = 11.33VIFSTVAHH475 pKa = 6.64SLTVIPSSADD485 pKa = 3.14TEE487 pKa = 4.46YY488 pKa = 11.73GLVIANGNNQDD499 pKa = 3.45YY500 pKa = 10.32QSRR503 pKa = 11.84VEE505 pKa = 4.79IDD507 pKa = 3.84DD508 pKa = 4.13DD509 pKa = 4.78ANYY512 pKa = 10.82LVIGSDD518 pKa = 3.07AKK520 pKa = 11.11DD521 pKa = 3.42IIVTGAGDD529 pKa = 4.0TIVSGGAGDD538 pKa = 4.97DD539 pKa = 4.83DD540 pKa = 3.7ITGGVGQDD548 pKa = 3.25IIFGGVGQDD557 pKa = 3.33HH558 pKa = 6.44VEE560 pKa = 4.29SGAGADD566 pKa = 3.53VVYY569 pKa = 10.79GGDD572 pKa = 4.5DD573 pKa = 3.6NDD575 pKa = 3.99TFIAQIGPMGLGGDD589 pKa = 3.95TYY591 pKa = 11.46YY592 pKa = 11.3GGDD595 pKa = 3.24ATSRR599 pKa = 11.84GSVDD603 pKa = 2.57GWDD606 pKa = 3.42VVDD609 pKa = 4.47YY610 pKa = 10.75RR611 pKa = 11.84QLDD614 pKa = 3.76TEE616 pKa = 4.71LYY618 pKa = 9.98GLAFEE623 pKa = 5.01QVDD626 pKa = 3.3AGHH629 pKa = 6.89RR630 pKa = 11.84RR631 pKa = 11.84LFMYY635 pKa = 9.59VKK637 pKa = 10.56NPGEE641 pKa = 4.03AQPLVFEE648 pKa = 5.23NYY650 pKa = 10.16DD651 pKa = 3.43DD652 pKa = 4.86LYY654 pKa = 10.93SIEE657 pKa = 5.24SILHH661 pKa = 5.69NDD663 pKa = 3.32YY664 pKa = 11.35LFDD667 pKa = 3.65VQFGDD672 pKa = 3.82EE673 pKa = 4.08VANNIRR679 pKa = 11.84AGDD682 pKa = 3.7SGGSGGGIVYY692 pKa = 10.15GRR694 pKa = 11.84GGNDD698 pKa = 3.27VLRR701 pKa = 11.84GASYY705 pKa = 8.36DD706 pKa = 3.57TNYY709 pKa = 10.97LFGEE713 pKa = 4.71DD714 pKa = 4.74GNDD717 pKa = 3.21TLYY720 pKa = 11.28SGDD723 pKa = 3.75AVNGSKK729 pKa = 10.7NFLWGGSGDD738 pKa = 3.76DD739 pKa = 3.7TLVGDD744 pKa = 3.89RR745 pKa = 11.84GEE747 pKa = 3.87NHH749 pKa = 7.45LYY751 pKa = 11.09GEE753 pKa = 4.55TGNDD757 pKa = 3.12TLTIANGGSGVYY769 pKa = 10.31DD770 pKa = 3.9GGADD774 pKa = 3.58FDD776 pKa = 4.64LVSGNLTNKK785 pKa = 9.39MYY787 pKa = 11.05LFADD791 pKa = 3.92TLSEE795 pKa = 4.15GGTYY799 pKa = 10.2TFVDD803 pKa = 3.23ADD805 pKa = 4.1GNNSLTVKK813 pKa = 10.47NMEE816 pKa = 4.2RR817 pKa = 11.84VGLTLSQTHH826 pKa = 6.22IAVTDD831 pKa = 3.62TQSFSNYY838 pKa = 9.2YY839 pKa = 8.59YY840 pKa = 11.37ANDD843 pKa = 3.95GEE845 pKa = 4.58FNYY848 pKa = 10.71LSLYY852 pKa = 10.02KK853 pKa = 10.61AIFDD857 pKa = 4.05FSGSEE862 pKa = 3.78TALTHH867 pKa = 6.08DD868 pKa = 3.68TAGYY872 pKa = 9.46GYY874 pKa = 11.09VNGVYY879 pKa = 9.98TLMDD883 pKa = 3.58YY884 pKa = 10.39EE885 pKa = 4.72VNGGSYY891 pKa = 7.43THH893 pKa = 6.27YY894 pKa = 8.83VQNVAGVVGSNMGDD908 pKa = 3.39TYY910 pKa = 11.67NLNSQQTTLYY920 pKa = 9.54MGTGDD925 pKa = 3.7DD926 pKa = 3.92TVNMGSRR933 pKa = 11.84TANDD937 pKa = 3.45VIYY940 pKa = 10.99SGGNDD945 pKa = 3.16VFYY948 pKa = 11.25SEE950 pKa = 5.49YY951 pKa = 9.27YY952 pKa = 10.63LPGITVDD959 pKa = 4.31AKK961 pKa = 10.88FLNTDD966 pKa = 2.98ATVTINSSLYY976 pKa = 10.92DD977 pKa = 3.58NGLSKK982 pKa = 11.08LDD984 pKa = 3.34ILFTFDD990 pKa = 4.14ADD992 pKa = 3.71NSIHH996 pKa = 6.7IIWEE1000 pKa = 4.31GSSYY1004 pKa = 10.85SASSSLKK1011 pKa = 9.75FYY1013 pKa = 10.72NATRR1017 pKa = 11.84IDD1019 pKa = 3.79FNGPQLMNPGSPDD1032 pKa = 3.38AYY1034 pKa = 10.57VKK1036 pKa = 10.46AAEE1039 pKa = 4.04YY1040 pKa = 10.58SLVGSTPVASTLEE1053 pKa = 4.18GTLGHH1058 pKa = 6.34DD1059 pKa = 4.1TIVSRR1064 pKa = 11.84SGYY1067 pKa = 8.56GQALIGLWGDD1077 pKa = 3.66DD1078 pKa = 3.55KK1079 pKa = 11.75LYY1081 pKa = 10.84GYY1083 pKa = 10.63DD1084 pKa = 4.96GADD1087 pKa = 3.45TLQGGAGDD1095 pKa = 3.75DD1096 pKa = 3.63TLYY1099 pKa = 11.22GGTGDD1104 pKa = 5.14DD1105 pKa = 4.15VLTGGADD1112 pKa = 3.15NDD1114 pKa = 3.65EE1115 pKa = 5.5FIFEE1119 pKa = 4.61SGFGHH1124 pKa = 6.95DD1125 pKa = 4.27TITDD1129 pKa = 4.13FVRR1132 pKa = 11.84GSDD1135 pKa = 3.78KK1136 pKa = 10.37ITFDD1140 pKa = 3.84SLSGIDD1146 pKa = 5.13DD1147 pKa = 3.73MSDD1150 pKa = 2.92LTFSAEE1156 pKa = 4.41TPTSTRR1162 pKa = 11.84IAVDD1166 pKa = 3.39SNNWVIVANQEE1177 pKa = 3.62FDD1179 pKa = 3.06SWLVSDD1185 pKa = 5.56FGFSTLLPVNGTSANDD1201 pKa = 3.45ILDD1204 pKa = 3.99GTSAAEE1210 pKa = 4.06EE1211 pKa = 3.99FHH1213 pKa = 7.16AGAGNDD1219 pKa = 3.57YY1220 pKa = 10.79VYY1222 pKa = 10.99AGAGNDD1228 pKa = 3.42VLYY1231 pKa = 11.23GEE1233 pKa = 5.57DD1234 pKa = 4.41GDD1236 pKa = 6.21DD1237 pKa = 3.35ILAGQFGNDD1246 pKa = 3.14TLYY1249 pKa = 11.2GGNGGDD1255 pKa = 3.64YY1256 pKa = 10.93LWGNEE1261 pKa = 4.53GNDD1264 pKa = 3.63TLYY1267 pKa = 11.33GGAGVDD1273 pKa = 3.4ALRR1276 pKa = 11.84GLEE1279 pKa = 4.42GDD1281 pKa = 5.04DD1282 pKa = 3.87ILHH1285 pKa = 6.83GGDD1288 pKa = 3.85GNDD1291 pKa = 3.45NLRR1294 pKa = 11.84GDD1296 pKa = 5.29DD1297 pKa = 3.76VDD1299 pKa = 4.74SDD1301 pKa = 4.08PSFAGAGNDD1310 pKa = 3.65ILYY1313 pKa = 10.78GDD1315 pKa = 5.01AGNDD1319 pKa = 3.49SLAGGAGNDD1328 pKa = 3.33ILRR1331 pKa = 11.84GGTGQDD1337 pKa = 3.8SLWGNAGSDD1346 pKa = 3.34TFLFMAEE1353 pKa = 4.4DD1354 pKa = 3.98LDD1356 pKa = 4.08GNRR1359 pKa = 11.84DD1360 pKa = 3.71YY1361 pKa = 10.93IYY1363 pKa = 10.79DD1364 pKa = 3.66FTVGSGGDD1372 pKa = 3.65MIDD1375 pKa = 3.3ISDD1378 pKa = 3.89VLSGYY1383 pKa = 10.95DD1384 pKa = 3.4SANDD1388 pKa = 3.45NLADD1392 pKa = 4.04FVSILTGGSYY1402 pKa = 9.82TRR1404 pKa = 11.84ISVDD1408 pKa = 2.79IDD1410 pKa = 3.59GTGTNYY1416 pKa = 10.39GSQSIIQIQGVTGLTLNDD1434 pKa = 3.35LTTNGNLIVEE1444 pKa = 4.82AAA1446 pKa = 3.69
MM1 pKa = 7.36TSDD4 pKa = 3.11ILMNAILSLDD14 pKa = 3.68AYY16 pKa = 10.7NRR18 pKa = 11.84GYY20 pKa = 10.87DD21 pKa = 3.2AGVQITEE28 pKa = 4.58PDD30 pKa = 4.52DD31 pKa = 3.16ITPYY35 pKa = 10.71KK36 pKa = 10.33VGNATFLKK44 pKa = 9.49QSSVAEE50 pKa = 4.07SSEE53 pKa = 3.88GVTAGFYY60 pKa = 10.67AIAYY64 pKa = 8.08SYY66 pKa = 11.23NGEE69 pKa = 4.23TVISYY74 pKa = 10.46RR75 pKa = 11.84GTDD78 pKa = 3.81DD79 pKa = 3.57YY80 pKa = 12.03DD81 pKa = 5.27GITSIPSSLDD91 pKa = 3.21AANGWPMGGGSITTAQGRR109 pKa = 11.84MALEE113 pKa = 4.82FYY115 pKa = 10.94KK116 pKa = 10.83DD117 pKa = 3.83VADD120 pKa = 4.48LPYY123 pKa = 10.94GSDD126 pKa = 3.38LRR128 pKa = 11.84NVAITLTGHH137 pKa = 5.85SLGGGLAGYY146 pKa = 8.81VASLYY151 pKa = 9.94GQNADD156 pKa = 4.3IFDD159 pKa = 3.78SMAFKK164 pKa = 10.68LAATATINVAYY175 pKa = 10.02HH176 pKa = 6.1PEE178 pKa = 4.48YY179 pKa = 10.15YY180 pKa = 9.84TDD182 pKa = 3.52PEE184 pKa = 4.74YY185 pKa = 11.09YY186 pKa = 9.7PEE188 pKa = 5.76PIFSDD193 pKa = 3.43AQEE196 pKa = 4.06LVYY199 pKa = 10.84NGYY202 pKa = 7.98STPWEE207 pKa = 4.19NSHH210 pKa = 6.85PEE212 pKa = 3.97YY213 pKa = 11.22VNGYY217 pKa = 8.02EE218 pKa = 5.33LSGQVISEE226 pKa = 4.38EE227 pKa = 3.69NWYY230 pKa = 6.7PTPGVDD236 pKa = 3.19SVGYY240 pKa = 8.25NTYY243 pKa = 11.17DD244 pKa = 5.27DD245 pKa = 5.41GDD247 pKa = 4.03LDD249 pKa = 5.25SNDD252 pKa = 3.46KK253 pKa = 11.03HH254 pKa = 8.04SMSSFVIRR262 pKa = 11.84YY263 pKa = 6.44YY264 pKa = 11.44AEE266 pKa = 3.71VGGANDD272 pKa = 4.01SALSTDD278 pKa = 3.35WQNAAPYY285 pKa = 8.7FWAVLYY291 pKa = 10.49DD292 pKa = 3.84DD293 pKa = 5.3QLAQAIGADD302 pKa = 3.84AKK304 pKa = 11.17DD305 pKa = 3.56GTLKK309 pKa = 11.11DD310 pKa = 3.34EE311 pKa = 4.51GKK313 pKa = 9.86YY314 pKa = 10.81AEE316 pKa = 4.24IVRR319 pKa = 11.84TAIAYY324 pKa = 9.66SSLEE328 pKa = 4.08KK329 pKa = 11.0NVDD332 pKa = 3.14GTGLIFGDD340 pKa = 3.35TGIRR344 pKa = 11.84ALYY347 pKa = 10.61DD348 pKa = 3.52DD349 pKa = 5.58ANDD352 pKa = 3.55LGARR356 pKa = 11.84IADD359 pKa = 4.16PLSGDD364 pKa = 3.86LFDD367 pKa = 4.42TYY369 pKa = 11.01GTVISRR375 pKa = 11.84AFVEE379 pKa = 4.95FAGSLALHH387 pKa = 6.84EE388 pKa = 4.23IEE390 pKa = 4.08KK391 pKa = 10.17TEE393 pKa = 3.83FAGAINGVLSHH404 pKa = 7.57DD405 pKa = 4.14ATNHH409 pKa = 5.21TLTVNYY415 pKa = 10.6ADD417 pKa = 4.87ALWIAAGKK425 pKa = 8.94GANDD429 pKa = 3.76IIVSRR434 pKa = 11.84EE435 pKa = 3.96NIVEE439 pKa = 3.97QAFGVDD445 pKa = 3.49EE446 pKa = 4.48TQLRR450 pKa = 11.84TAMWDD455 pKa = 3.03LWQNDD460 pKa = 3.28GNAVFDD466 pKa = 3.87KK467 pKa = 11.33VIFSTVAHH475 pKa = 6.64SLTVIPSSADD485 pKa = 3.14TEE487 pKa = 4.46YY488 pKa = 11.73GLVIANGNNQDD499 pKa = 3.45YY500 pKa = 10.32QSRR503 pKa = 11.84VEE505 pKa = 4.79IDD507 pKa = 3.84DD508 pKa = 4.13DD509 pKa = 4.78ANYY512 pKa = 10.82LVIGSDD518 pKa = 3.07AKK520 pKa = 11.11DD521 pKa = 3.42IIVTGAGDD529 pKa = 4.0TIVSGGAGDD538 pKa = 4.97DD539 pKa = 4.83DD540 pKa = 3.7ITGGVGQDD548 pKa = 3.25IIFGGVGQDD557 pKa = 3.33HH558 pKa = 6.44VEE560 pKa = 4.29SGAGADD566 pKa = 3.53VVYY569 pKa = 10.79GGDD572 pKa = 4.5DD573 pKa = 3.6NDD575 pKa = 3.99TFIAQIGPMGLGGDD589 pKa = 3.95TYY591 pKa = 11.46YY592 pKa = 11.3GGDD595 pKa = 3.24ATSRR599 pKa = 11.84GSVDD603 pKa = 2.57GWDD606 pKa = 3.42VVDD609 pKa = 4.47YY610 pKa = 10.75RR611 pKa = 11.84QLDD614 pKa = 3.76TEE616 pKa = 4.71LYY618 pKa = 9.98GLAFEE623 pKa = 5.01QVDD626 pKa = 3.3AGHH629 pKa = 6.89RR630 pKa = 11.84RR631 pKa = 11.84LFMYY635 pKa = 9.59VKK637 pKa = 10.56NPGEE641 pKa = 4.03AQPLVFEE648 pKa = 5.23NYY650 pKa = 10.16DD651 pKa = 3.43DD652 pKa = 4.86LYY654 pKa = 10.93SIEE657 pKa = 5.24SILHH661 pKa = 5.69NDD663 pKa = 3.32YY664 pKa = 11.35LFDD667 pKa = 3.65VQFGDD672 pKa = 3.82EE673 pKa = 4.08VANNIRR679 pKa = 11.84AGDD682 pKa = 3.7SGGSGGGIVYY692 pKa = 10.15GRR694 pKa = 11.84GGNDD698 pKa = 3.27VLRR701 pKa = 11.84GASYY705 pKa = 8.36DD706 pKa = 3.57TNYY709 pKa = 10.97LFGEE713 pKa = 4.71DD714 pKa = 4.74GNDD717 pKa = 3.21TLYY720 pKa = 11.28SGDD723 pKa = 3.75AVNGSKK729 pKa = 10.7NFLWGGSGDD738 pKa = 3.76DD739 pKa = 3.7TLVGDD744 pKa = 3.89RR745 pKa = 11.84GEE747 pKa = 3.87NHH749 pKa = 7.45LYY751 pKa = 11.09GEE753 pKa = 4.55TGNDD757 pKa = 3.12TLTIANGGSGVYY769 pKa = 10.31DD770 pKa = 3.9GGADD774 pKa = 3.58FDD776 pKa = 4.64LVSGNLTNKK785 pKa = 9.39MYY787 pKa = 11.05LFADD791 pKa = 3.92TLSEE795 pKa = 4.15GGTYY799 pKa = 10.2TFVDD803 pKa = 3.23ADD805 pKa = 4.1GNNSLTVKK813 pKa = 10.47NMEE816 pKa = 4.2RR817 pKa = 11.84VGLTLSQTHH826 pKa = 6.22IAVTDD831 pKa = 3.62TQSFSNYY838 pKa = 9.2YY839 pKa = 8.59YY840 pKa = 11.37ANDD843 pKa = 3.95GEE845 pKa = 4.58FNYY848 pKa = 10.71LSLYY852 pKa = 10.02KK853 pKa = 10.61AIFDD857 pKa = 4.05FSGSEE862 pKa = 3.78TALTHH867 pKa = 6.08DD868 pKa = 3.68TAGYY872 pKa = 9.46GYY874 pKa = 11.09VNGVYY879 pKa = 9.98TLMDD883 pKa = 3.58YY884 pKa = 10.39EE885 pKa = 4.72VNGGSYY891 pKa = 7.43THH893 pKa = 6.27YY894 pKa = 8.83VQNVAGVVGSNMGDD908 pKa = 3.39TYY910 pKa = 11.67NLNSQQTTLYY920 pKa = 9.54MGTGDD925 pKa = 3.7DD926 pKa = 3.92TVNMGSRR933 pKa = 11.84TANDD937 pKa = 3.45VIYY940 pKa = 10.99SGGNDD945 pKa = 3.16VFYY948 pKa = 11.25SEE950 pKa = 5.49YY951 pKa = 9.27YY952 pKa = 10.63LPGITVDD959 pKa = 4.31AKK961 pKa = 10.88FLNTDD966 pKa = 2.98ATVTINSSLYY976 pKa = 10.92DD977 pKa = 3.58NGLSKK982 pKa = 11.08LDD984 pKa = 3.34ILFTFDD990 pKa = 4.14ADD992 pKa = 3.71NSIHH996 pKa = 6.7IIWEE1000 pKa = 4.31GSSYY1004 pKa = 10.85SASSSLKK1011 pKa = 9.75FYY1013 pKa = 10.72NATRR1017 pKa = 11.84IDD1019 pKa = 3.79FNGPQLMNPGSPDD1032 pKa = 3.38AYY1034 pKa = 10.57VKK1036 pKa = 10.46AAEE1039 pKa = 4.04YY1040 pKa = 10.58SLVGSTPVASTLEE1053 pKa = 4.18GTLGHH1058 pKa = 6.34DD1059 pKa = 4.1TIVSRR1064 pKa = 11.84SGYY1067 pKa = 8.56GQALIGLWGDD1077 pKa = 3.66DD1078 pKa = 3.55KK1079 pKa = 11.75LYY1081 pKa = 10.84GYY1083 pKa = 10.63DD1084 pKa = 4.96GADD1087 pKa = 3.45TLQGGAGDD1095 pKa = 3.75DD1096 pKa = 3.63TLYY1099 pKa = 11.22GGTGDD1104 pKa = 5.14DD1105 pKa = 4.15VLTGGADD1112 pKa = 3.15NDD1114 pKa = 3.65EE1115 pKa = 5.5FIFEE1119 pKa = 4.61SGFGHH1124 pKa = 6.95DD1125 pKa = 4.27TITDD1129 pKa = 4.13FVRR1132 pKa = 11.84GSDD1135 pKa = 3.78KK1136 pKa = 10.37ITFDD1140 pKa = 3.84SLSGIDD1146 pKa = 5.13DD1147 pKa = 3.73MSDD1150 pKa = 2.92LTFSAEE1156 pKa = 4.41TPTSTRR1162 pKa = 11.84IAVDD1166 pKa = 3.39SNNWVIVANQEE1177 pKa = 3.62FDD1179 pKa = 3.06SWLVSDD1185 pKa = 5.56FGFSTLLPVNGTSANDD1201 pKa = 3.45ILDD1204 pKa = 3.99GTSAAEE1210 pKa = 4.06EE1211 pKa = 3.99FHH1213 pKa = 7.16AGAGNDD1219 pKa = 3.57YY1220 pKa = 10.79VYY1222 pKa = 10.99AGAGNDD1228 pKa = 3.42VLYY1231 pKa = 11.23GEE1233 pKa = 5.57DD1234 pKa = 4.41GDD1236 pKa = 6.21DD1237 pKa = 3.35ILAGQFGNDD1246 pKa = 3.14TLYY1249 pKa = 11.2GGNGGDD1255 pKa = 3.64YY1256 pKa = 10.93LWGNEE1261 pKa = 4.53GNDD1264 pKa = 3.63TLYY1267 pKa = 11.33GGAGVDD1273 pKa = 3.4ALRR1276 pKa = 11.84GLEE1279 pKa = 4.42GDD1281 pKa = 5.04DD1282 pKa = 3.87ILHH1285 pKa = 6.83GGDD1288 pKa = 3.85GNDD1291 pKa = 3.45NLRR1294 pKa = 11.84GDD1296 pKa = 5.29DD1297 pKa = 3.76VDD1299 pKa = 4.74SDD1301 pKa = 4.08PSFAGAGNDD1310 pKa = 3.65ILYY1313 pKa = 10.78GDD1315 pKa = 5.01AGNDD1319 pKa = 3.49SLAGGAGNDD1328 pKa = 3.33ILRR1331 pKa = 11.84GGTGQDD1337 pKa = 3.8SLWGNAGSDD1346 pKa = 3.34TFLFMAEE1353 pKa = 4.4DD1354 pKa = 3.98LDD1356 pKa = 4.08GNRR1359 pKa = 11.84DD1360 pKa = 3.71YY1361 pKa = 10.93IYY1363 pKa = 10.79DD1364 pKa = 3.66FTVGSGGDD1372 pKa = 3.65MIDD1375 pKa = 3.3ISDD1378 pKa = 3.89VLSGYY1383 pKa = 10.95DD1384 pKa = 3.4SANDD1388 pKa = 3.45NLADD1392 pKa = 4.04FVSILTGGSYY1402 pKa = 9.82TRR1404 pKa = 11.84ISVDD1408 pKa = 2.79IDD1410 pKa = 3.59GTGTNYY1416 pKa = 10.39GSQSIIQIQGVTGLTLNDD1434 pKa = 3.35LTTNGNLIVEE1444 pKa = 4.82AAA1446 pKa = 3.69
Molecular weight: 153.52 kDa
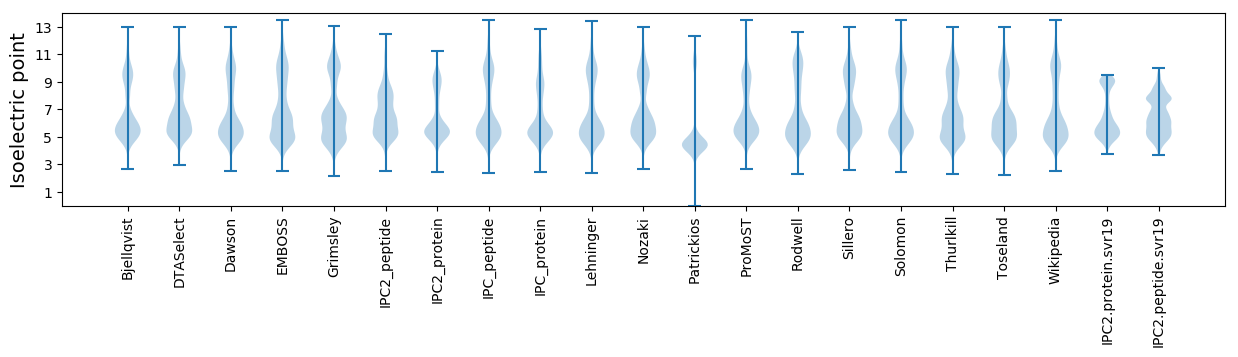
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A239EUW1|A0A239EUW1_9SPHN 3-hydroxyisobutyrate dehydrogenase OS=Sphingomonas laterariae OX=861865 GN=SAMN06295912_107112 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.61GFRR19 pKa = 11.84SRR21 pKa = 11.84SATPGGRR28 pKa = 11.84KK29 pKa = 9.04VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 8.96RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.61GFRR19 pKa = 11.84SRR21 pKa = 11.84SATPGGRR28 pKa = 11.84KK29 pKa = 9.04VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1329728 |
27 |
4406 |
311.0 |
33.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.033 ± 0.065 | 0.751 ± 0.011 |
6.17 ± 0.03 | 5.334 ± 0.04 |
3.496 ± 0.025 | 9.019 ± 0.062 |
1.998 ± 0.019 | 5.168 ± 0.028 |
2.878 ± 0.028 | 9.725 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.402 ± 0.021 | 2.454 ± 0.024 |
5.376 ± 0.033 | 3.005 ± 0.021 |
7.568 ± 0.044 | 4.881 ± 0.027 |
5.141 ± 0.038 | 6.994 ± 0.033 |
1.44 ± 0.017 | 2.166 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
