
Trichomonas vaginalis virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Trichomonasvirus
Average proteome isoelectric point is 7.43
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q9JE95|Q9JE95_9VIRU Capsid protein OS=Trichomonas vaginalis virus 2 OX=674954 GN=cap PE=4 SV=1
MM1 pKa = 7.93ASTLISSDD9 pKa = 3.08NSATLGKK16 pKa = 10.35DD17 pKa = 3.02SEE19 pKa = 4.95VINNTNTSPPDD30 pKa = 3.64NPSSDD35 pKa = 3.19HH36 pKa = 6.59SNPRR40 pKa = 11.84LTKK43 pKa = 10.68VLDD46 pKa = 3.76EE47 pKa = 4.31MSKK50 pKa = 10.5KK51 pKa = 10.01PCVNINEE58 pKa = 3.97IRR60 pKa = 11.84KK61 pKa = 8.57MIRR64 pKa = 11.84NFQPQFIQPRR74 pKa = 11.84NGNRR78 pKa = 11.84PNAQPRR84 pKa = 11.84TVDD87 pKa = 3.01SFEE90 pKa = 3.93WVVRR94 pKa = 11.84IQSTVEE100 pKa = 3.96TQLLGATNTVPQQTLNLDD118 pKa = 3.11ISFTDD123 pKa = 3.96DD124 pKa = 3.09STTITPASIPGSISMLDD141 pKa = 3.55NSRR144 pKa = 11.84HH145 pKa = 5.08IPAIQSMIQNFKK157 pKa = 10.91ARR159 pKa = 11.84YY160 pKa = 8.88LGSLQDD166 pKa = 3.7TAQLQSPQYY175 pKa = 8.61PQLLAYY181 pKa = 10.86LFGQLIAIKK190 pKa = 10.48DD191 pKa = 3.73RR192 pKa = 11.84LDD194 pKa = 3.27LFRR197 pKa = 11.84PSNPLSLADD206 pKa = 3.75ALFGFTLAQNARR218 pKa = 11.84PRR220 pKa = 11.84YY221 pKa = 9.04DD222 pKa = 3.31DD223 pKa = 3.73HH224 pKa = 8.01RR225 pKa = 11.84HH226 pKa = 5.62AKK228 pKa = 9.39ACQGPLVIPAATNSDD243 pKa = 4.07CGPCGFVQINANQGLTLPLGACLFVNPEE271 pKa = 4.19TVNDD275 pKa = 3.5QSFQDD280 pKa = 3.96FLWLIFATHH289 pKa = 6.09HH290 pKa = 6.33RR291 pKa = 11.84MPNQMQNNWPFSLNIVSTCAAPGRR315 pKa = 11.84QAPHH319 pKa = 6.56AGEE322 pKa = 4.11LTDD325 pKa = 3.43EE326 pKa = 4.34RR327 pKa = 11.84VRR329 pKa = 11.84LALDD333 pKa = 3.26TGHH336 pKa = 7.49RR337 pKa = 11.84ILLSMFNDD345 pKa = 3.74DD346 pKa = 4.95EE347 pKa = 4.42EE348 pKa = 4.43TLRR351 pKa = 11.84YY352 pKa = 8.24YY353 pKa = 9.44QRR355 pKa = 11.84KK356 pKa = 9.4GIEE359 pKa = 4.05TMFRR363 pKa = 11.84PCCFYY368 pKa = 11.09TEE370 pKa = 4.28GGLLRR375 pKa = 11.84KK376 pKa = 8.05ATRR379 pKa = 11.84YY380 pKa = 10.52VSMVPLNGLYY390 pKa = 10.11YY391 pKa = 10.4YY392 pKa = 10.82NGATSYY398 pKa = 10.68VVSPIHH404 pKa = 6.46TDD406 pKa = 2.87AHH408 pKa = 6.09PGITAAIEE416 pKa = 4.22SFVDD420 pKa = 3.44IMVLQAVFSFSGPKK434 pKa = 9.47VVAAKK439 pKa = 10.65VNASQIDD446 pKa = 3.45AAMVFGPAVAEE457 pKa = 4.16GDD459 pKa = 3.77GFVYY463 pKa = 10.54DD464 pKa = 4.78PLRR467 pKa = 11.84PAPPLSAFYY476 pKa = 10.67TEE478 pKa = 5.78FIHH481 pKa = 7.35RR482 pKa = 11.84PAEE485 pKa = 3.77QRR487 pKa = 11.84IFQMAMSQIYY497 pKa = 10.02GSHH500 pKa = 6.77APLIIANVINSIHH513 pKa = 5.59NCKK516 pKa = 9.21TKK518 pKa = 10.35IVNNKK523 pKa = 8.71LRR525 pKa = 11.84ATFVRR530 pKa = 11.84RR531 pKa = 11.84PPGAPHH537 pKa = 7.04LKK539 pKa = 10.38ADD541 pKa = 3.58TAIINRR547 pKa = 11.84FHH549 pKa = 7.29DD550 pKa = 4.02PEE552 pKa = 4.14LAYY555 pKa = 10.6ALGILADD562 pKa = 5.49GIAPLDD568 pKa = 3.94GSHH571 pKa = 6.94EE572 pKa = 4.26YY573 pKa = 10.78NVLDD577 pKa = 3.95EE578 pKa = 5.65LDD580 pKa = 3.58YY581 pKa = 11.49LFNGGDD587 pKa = 3.14IRR589 pKa = 11.84NCFGLNALNTRR600 pKa = 11.84GLGQIVHH607 pKa = 6.63IRR609 pKa = 11.84PKK611 pKa = 10.19RR612 pKa = 11.84EE613 pKa = 3.37PGKK616 pKa = 9.46RR617 pKa = 11.84PRR619 pKa = 11.84RR620 pKa = 11.84GFYY623 pKa = 7.56TTLDD627 pKa = 3.99GQVHH631 pKa = 7.0PVTQDD636 pKa = 2.83APLDD640 pKa = 4.48EE641 pKa = 5.1IYY643 pKa = 10.24HH644 pKa = 5.68WRR646 pKa = 11.84DD647 pKa = 2.85HH648 pKa = 6.76GNLTRR653 pKa = 11.84PYY655 pKa = 9.27SCHH658 pKa = 7.05ILDD661 pKa = 3.83SQGLEE666 pKa = 3.99FADD669 pKa = 3.66VSNGRR674 pKa = 11.84SRR676 pKa = 11.84GKK678 pKa = 9.94ILVVVNSPLKK688 pKa = 9.97TCAAYY693 pKa = 9.98QGPSFAPKK701 pKa = 9.62PGSAMWNEE709 pKa = 4.0
MM1 pKa = 7.93ASTLISSDD9 pKa = 3.08NSATLGKK16 pKa = 10.35DD17 pKa = 3.02SEE19 pKa = 4.95VINNTNTSPPDD30 pKa = 3.64NPSSDD35 pKa = 3.19HH36 pKa = 6.59SNPRR40 pKa = 11.84LTKK43 pKa = 10.68VLDD46 pKa = 3.76EE47 pKa = 4.31MSKK50 pKa = 10.5KK51 pKa = 10.01PCVNINEE58 pKa = 3.97IRR60 pKa = 11.84KK61 pKa = 8.57MIRR64 pKa = 11.84NFQPQFIQPRR74 pKa = 11.84NGNRR78 pKa = 11.84PNAQPRR84 pKa = 11.84TVDD87 pKa = 3.01SFEE90 pKa = 3.93WVVRR94 pKa = 11.84IQSTVEE100 pKa = 3.96TQLLGATNTVPQQTLNLDD118 pKa = 3.11ISFTDD123 pKa = 3.96DD124 pKa = 3.09STTITPASIPGSISMLDD141 pKa = 3.55NSRR144 pKa = 11.84HH145 pKa = 5.08IPAIQSMIQNFKK157 pKa = 10.91ARR159 pKa = 11.84YY160 pKa = 8.88LGSLQDD166 pKa = 3.7TAQLQSPQYY175 pKa = 8.61PQLLAYY181 pKa = 10.86LFGQLIAIKK190 pKa = 10.48DD191 pKa = 3.73RR192 pKa = 11.84LDD194 pKa = 3.27LFRR197 pKa = 11.84PSNPLSLADD206 pKa = 3.75ALFGFTLAQNARR218 pKa = 11.84PRR220 pKa = 11.84YY221 pKa = 9.04DD222 pKa = 3.31DD223 pKa = 3.73HH224 pKa = 8.01RR225 pKa = 11.84HH226 pKa = 5.62AKK228 pKa = 9.39ACQGPLVIPAATNSDD243 pKa = 4.07CGPCGFVQINANQGLTLPLGACLFVNPEE271 pKa = 4.19TVNDD275 pKa = 3.5QSFQDD280 pKa = 3.96FLWLIFATHH289 pKa = 6.09HH290 pKa = 6.33RR291 pKa = 11.84MPNQMQNNWPFSLNIVSTCAAPGRR315 pKa = 11.84QAPHH319 pKa = 6.56AGEE322 pKa = 4.11LTDD325 pKa = 3.43EE326 pKa = 4.34RR327 pKa = 11.84VRR329 pKa = 11.84LALDD333 pKa = 3.26TGHH336 pKa = 7.49RR337 pKa = 11.84ILLSMFNDD345 pKa = 3.74DD346 pKa = 4.95EE347 pKa = 4.42EE348 pKa = 4.43TLRR351 pKa = 11.84YY352 pKa = 8.24YY353 pKa = 9.44QRR355 pKa = 11.84KK356 pKa = 9.4GIEE359 pKa = 4.05TMFRR363 pKa = 11.84PCCFYY368 pKa = 11.09TEE370 pKa = 4.28GGLLRR375 pKa = 11.84KK376 pKa = 8.05ATRR379 pKa = 11.84YY380 pKa = 10.52VSMVPLNGLYY390 pKa = 10.11YY391 pKa = 10.4YY392 pKa = 10.82NGATSYY398 pKa = 10.68VVSPIHH404 pKa = 6.46TDD406 pKa = 2.87AHH408 pKa = 6.09PGITAAIEE416 pKa = 4.22SFVDD420 pKa = 3.44IMVLQAVFSFSGPKK434 pKa = 9.47VVAAKK439 pKa = 10.65VNASQIDD446 pKa = 3.45AAMVFGPAVAEE457 pKa = 4.16GDD459 pKa = 3.77GFVYY463 pKa = 10.54DD464 pKa = 4.78PLRR467 pKa = 11.84PAPPLSAFYY476 pKa = 10.67TEE478 pKa = 5.78FIHH481 pKa = 7.35RR482 pKa = 11.84PAEE485 pKa = 3.77QRR487 pKa = 11.84IFQMAMSQIYY497 pKa = 10.02GSHH500 pKa = 6.77APLIIANVINSIHH513 pKa = 5.59NCKK516 pKa = 9.21TKK518 pKa = 10.35IVNNKK523 pKa = 8.71LRR525 pKa = 11.84ATFVRR530 pKa = 11.84RR531 pKa = 11.84PPGAPHH537 pKa = 7.04LKK539 pKa = 10.38ADD541 pKa = 3.58TAIINRR547 pKa = 11.84FHH549 pKa = 7.29DD550 pKa = 4.02PEE552 pKa = 4.14LAYY555 pKa = 10.6ALGILADD562 pKa = 5.49GIAPLDD568 pKa = 3.94GSHH571 pKa = 6.94EE572 pKa = 4.26YY573 pKa = 10.78NVLDD577 pKa = 3.95EE578 pKa = 5.65LDD580 pKa = 3.58YY581 pKa = 11.49LFNGGDD587 pKa = 3.14IRR589 pKa = 11.84NCFGLNALNTRR600 pKa = 11.84GLGQIVHH607 pKa = 6.63IRR609 pKa = 11.84PKK611 pKa = 10.19RR612 pKa = 11.84EE613 pKa = 3.37PGKK616 pKa = 9.46RR617 pKa = 11.84PRR619 pKa = 11.84RR620 pKa = 11.84GFYY623 pKa = 7.56TTLDD627 pKa = 3.99GQVHH631 pKa = 7.0PVTQDD636 pKa = 2.83APLDD640 pKa = 4.48EE641 pKa = 5.1IYY643 pKa = 10.24HH644 pKa = 5.68WRR646 pKa = 11.84DD647 pKa = 2.85HH648 pKa = 6.76GNLTRR653 pKa = 11.84PYY655 pKa = 9.27SCHH658 pKa = 7.05ILDD661 pKa = 3.83SQGLEE666 pKa = 3.99FADD669 pKa = 3.66VSNGRR674 pKa = 11.84SRR676 pKa = 11.84GKK678 pKa = 9.94ILVVVNSPLKK688 pKa = 9.97TCAAYY693 pKa = 9.98QGPSFAPKK701 pKa = 9.62PGSAMWNEE709 pKa = 4.0
Molecular weight: 78.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9JE95|Q9JE95_9VIRU Capsid protein OS=Trichomonas vaginalis virus 2 OX=674954 GN=cap PE=4 SV=1
RR1 pKa = 6.96FQRR4 pKa = 11.84TVTRR8 pKa = 11.84KK9 pKa = 10.17DD10 pKa = 2.97PCGRR14 pKa = 11.84QLTPQDD20 pKa = 3.94MRR22 pKa = 11.84CLPGPQLRR30 pKa = 11.84AKK32 pKa = 10.21AGQRR36 pKa = 11.84HH37 pKa = 4.89VEE39 pKa = 4.54RR40 pKa = 11.84INQCGGSVIYY50 pKa = 9.88PRR52 pKa = 11.84LSTARR57 pKa = 11.84ACSLSAIDD65 pKa = 5.98KK66 pKa = 7.35MTLALAHH73 pKa = 5.55QLCYY77 pKa = 10.25LYY79 pKa = 10.69KK80 pKa = 10.67SSDD83 pKa = 3.19LHH85 pKa = 6.17RR86 pKa = 11.84QLDD89 pKa = 3.77TMIPQSYY96 pKa = 8.0LTFLEE101 pKa = 3.91WLLRR105 pKa = 11.84IDD107 pKa = 3.48PHH109 pKa = 6.93NEE111 pKa = 3.42KK112 pKa = 10.74SSIRR116 pKa = 11.84HH117 pKa = 5.58FPSQDD122 pKa = 2.63NHH124 pKa = 5.29EE125 pKa = 5.0VITHH129 pKa = 5.65SLRR132 pKa = 11.84NLTKK136 pKa = 9.6EE137 pKa = 3.83QEE139 pKa = 3.93ITLFPIKK146 pKa = 10.52DD147 pKa = 2.98IVQANRR153 pKa = 11.84RR154 pKa = 11.84VNAYY158 pKa = 9.82ARR160 pKa = 11.84NLLDD164 pKa = 5.15ASPLPDD170 pKa = 3.77FALQQMLLPNTANDD184 pKa = 3.8VVCAILLLGEE194 pKa = 4.42VLWMLRR200 pKa = 11.84CPISIIVNISRR211 pKa = 11.84AICRR215 pKa = 11.84NDD217 pKa = 3.35SFLKK221 pKa = 10.74DD222 pKa = 3.43LSDD225 pKa = 3.84FNKK228 pKa = 9.93MLGLTKK234 pKa = 10.12IPIANCLTEE243 pKa = 5.51LNTLQGRR250 pKa = 11.84GVTSSDD256 pKa = 2.82AKK258 pKa = 10.61RR259 pKa = 11.84DD260 pKa = 3.64LTHH263 pKa = 7.56RR264 pKa = 11.84IADD267 pKa = 4.02VNPHH271 pKa = 5.22EE272 pKa = 4.8AKK274 pKa = 10.25ISRR277 pKa = 11.84EE278 pKa = 3.85NLRR281 pKa = 11.84EE282 pKa = 4.51AINQIYY288 pKa = 9.9KK289 pKa = 10.78EE290 pKa = 4.17EE291 pKa = 4.08ITRR294 pKa = 11.84KK295 pKa = 9.16EE296 pKa = 4.0VPDD299 pKa = 3.52TFKK302 pKa = 10.9QHH304 pKa = 6.13VFTSPLWVKK313 pKa = 10.53KK314 pKa = 9.87GAHH317 pKa = 5.49HH318 pKa = 6.72HH319 pKa = 6.49PLFGSYY325 pKa = 10.84DD326 pKa = 3.38NRR328 pKa = 11.84LEE330 pKa = 3.97FVEE333 pKa = 5.31NVDD336 pKa = 3.68LDD338 pKa = 4.13RR339 pKa = 11.84VLKK342 pKa = 9.91SHH344 pKa = 6.76PAVYY348 pKa = 8.77ITQAPKK354 pKa = 10.64LEE356 pKa = 4.34HH357 pKa = 6.1GKK359 pKa = 8.01TRR361 pKa = 11.84FIYY364 pKa = 10.5NCDD367 pKa = 3.39TVSYY371 pKa = 10.35IYY373 pKa = 10.47FDD375 pKa = 4.16YY376 pKa = 10.49ILNYY380 pKa = 9.58IEE382 pKa = 5.32SVWSNKK388 pKa = 9.02HH389 pKa = 5.07VLLNPDD395 pKa = 3.29YY396 pKa = 9.06MNPVIFSTLNYY407 pKa = 10.36DD408 pKa = 4.33EE409 pKa = 5.48YY410 pKa = 11.96CMLDD414 pKa = 3.32YY415 pKa = 11.32TDD417 pKa = 6.17FNSQHH422 pKa = 6.72SIEE425 pKa = 4.36SMKK428 pKa = 10.7QVFLCLFPFLPRR440 pKa = 11.84SMHH443 pKa = 7.42SILQWCVTSFDD454 pKa = 3.3NMYY457 pKa = 10.56INKK460 pKa = 7.62THH462 pKa = 6.8WNSTLPSGHH471 pKa = 7.23RR472 pKa = 11.84ATTFINSVLNRR483 pKa = 11.84AYY485 pKa = 10.82LLPFLQVANAFHH497 pKa = 6.54TGDD500 pKa = 4.89DD501 pKa = 3.56VLLCGKK507 pKa = 10.2ADD509 pKa = 3.63YY510 pKa = 7.84ATLINTVPYY519 pKa = 9.77EE520 pKa = 3.98LNKK523 pKa = 9.5TKK525 pKa = 10.72QSFGPSAEE533 pKa = 3.75FLRR536 pKa = 11.84LHH538 pKa = 6.04KK539 pKa = 10.65HH540 pKa = 5.41NDD542 pKa = 3.25QVSGYY547 pKa = 7.28PARR550 pKa = 11.84AISSLVSGNWLSFANPLWQPSLLSIMQQLYY580 pKa = 9.42TISARR585 pKa = 11.84SGLLPYY591 pKa = 10.26IPVTMKK597 pKa = 11.03LEE599 pKa = 3.95VQRR602 pKa = 11.84RR603 pKa = 11.84YY604 pKa = 10.8DD605 pKa = 3.39LRR607 pKa = 11.84SRR609 pKa = 11.84ITNGLFSGDD618 pKa = 3.96IVPSGCPCYY627 pKa = 10.27KK628 pKa = 10.65SNAALLSAVVPDD640 pKa = 4.35TVVKK644 pKa = 10.68APPTFYY650 pKa = 11.06DD651 pKa = 4.84LRR653 pKa = 11.84TLDD656 pKa = 3.59ILKK659 pKa = 8.41QTSPWINSASRR670 pKa = 11.84YY671 pKa = 8.72MDD673 pKa = 3.86LLEE676 pKa = 4.28RR677 pKa = 11.84RR678 pKa = 11.84HH679 pKa = 5.83MEE681 pKa = 3.87SDD683 pKa = 3.3NKK685 pKa = 10.36NVIYY689 pKa = 10.28SIQYY693 pKa = 9.77LPSKK697 pKa = 8.38MLPMIDD703 pKa = 4.33VDD705 pKa = 4.23PADD708 pKa = 3.84ATPLRR713 pKa = 11.84KK714 pKa = 9.04RR715 pKa = 11.84YY716 pKa = 9.37HH717 pKa = 5.94PRR719 pKa = 11.84SHH721 pKa = 7.02IAHH724 pKa = 7.37PLPRR728 pKa = 11.84DD729 pKa = 3.3AHH731 pKa = 5.86LKK733 pKa = 8.18EE734 pKa = 4.81LRR736 pKa = 11.84FATCRR741 pKa = 11.84VGPATAIRR749 pKa = 11.84LGSLWPANRR758 pKa = 11.84INLIKK763 pKa = 10.1PVYY766 pKa = 9.27VV767 pKa = 3.37
RR1 pKa = 6.96FQRR4 pKa = 11.84TVTRR8 pKa = 11.84KK9 pKa = 10.17DD10 pKa = 2.97PCGRR14 pKa = 11.84QLTPQDD20 pKa = 3.94MRR22 pKa = 11.84CLPGPQLRR30 pKa = 11.84AKK32 pKa = 10.21AGQRR36 pKa = 11.84HH37 pKa = 4.89VEE39 pKa = 4.54RR40 pKa = 11.84INQCGGSVIYY50 pKa = 9.88PRR52 pKa = 11.84LSTARR57 pKa = 11.84ACSLSAIDD65 pKa = 5.98KK66 pKa = 7.35MTLALAHH73 pKa = 5.55QLCYY77 pKa = 10.25LYY79 pKa = 10.69KK80 pKa = 10.67SSDD83 pKa = 3.19LHH85 pKa = 6.17RR86 pKa = 11.84QLDD89 pKa = 3.77TMIPQSYY96 pKa = 8.0LTFLEE101 pKa = 3.91WLLRR105 pKa = 11.84IDD107 pKa = 3.48PHH109 pKa = 6.93NEE111 pKa = 3.42KK112 pKa = 10.74SSIRR116 pKa = 11.84HH117 pKa = 5.58FPSQDD122 pKa = 2.63NHH124 pKa = 5.29EE125 pKa = 5.0VITHH129 pKa = 5.65SLRR132 pKa = 11.84NLTKK136 pKa = 9.6EE137 pKa = 3.83QEE139 pKa = 3.93ITLFPIKK146 pKa = 10.52DD147 pKa = 2.98IVQANRR153 pKa = 11.84RR154 pKa = 11.84VNAYY158 pKa = 9.82ARR160 pKa = 11.84NLLDD164 pKa = 5.15ASPLPDD170 pKa = 3.77FALQQMLLPNTANDD184 pKa = 3.8VVCAILLLGEE194 pKa = 4.42VLWMLRR200 pKa = 11.84CPISIIVNISRR211 pKa = 11.84AICRR215 pKa = 11.84NDD217 pKa = 3.35SFLKK221 pKa = 10.74DD222 pKa = 3.43LSDD225 pKa = 3.84FNKK228 pKa = 9.93MLGLTKK234 pKa = 10.12IPIANCLTEE243 pKa = 5.51LNTLQGRR250 pKa = 11.84GVTSSDD256 pKa = 2.82AKK258 pKa = 10.61RR259 pKa = 11.84DD260 pKa = 3.64LTHH263 pKa = 7.56RR264 pKa = 11.84IADD267 pKa = 4.02VNPHH271 pKa = 5.22EE272 pKa = 4.8AKK274 pKa = 10.25ISRR277 pKa = 11.84EE278 pKa = 3.85NLRR281 pKa = 11.84EE282 pKa = 4.51AINQIYY288 pKa = 9.9KK289 pKa = 10.78EE290 pKa = 4.17EE291 pKa = 4.08ITRR294 pKa = 11.84KK295 pKa = 9.16EE296 pKa = 4.0VPDD299 pKa = 3.52TFKK302 pKa = 10.9QHH304 pKa = 6.13VFTSPLWVKK313 pKa = 10.53KK314 pKa = 9.87GAHH317 pKa = 5.49HH318 pKa = 6.72HH319 pKa = 6.49PLFGSYY325 pKa = 10.84DD326 pKa = 3.38NRR328 pKa = 11.84LEE330 pKa = 3.97FVEE333 pKa = 5.31NVDD336 pKa = 3.68LDD338 pKa = 4.13RR339 pKa = 11.84VLKK342 pKa = 9.91SHH344 pKa = 6.76PAVYY348 pKa = 8.77ITQAPKK354 pKa = 10.64LEE356 pKa = 4.34HH357 pKa = 6.1GKK359 pKa = 8.01TRR361 pKa = 11.84FIYY364 pKa = 10.5NCDD367 pKa = 3.39TVSYY371 pKa = 10.35IYY373 pKa = 10.47FDD375 pKa = 4.16YY376 pKa = 10.49ILNYY380 pKa = 9.58IEE382 pKa = 5.32SVWSNKK388 pKa = 9.02HH389 pKa = 5.07VLLNPDD395 pKa = 3.29YY396 pKa = 9.06MNPVIFSTLNYY407 pKa = 10.36DD408 pKa = 4.33EE409 pKa = 5.48YY410 pKa = 11.96CMLDD414 pKa = 3.32YY415 pKa = 11.32TDD417 pKa = 6.17FNSQHH422 pKa = 6.72SIEE425 pKa = 4.36SMKK428 pKa = 10.7QVFLCLFPFLPRR440 pKa = 11.84SMHH443 pKa = 7.42SILQWCVTSFDD454 pKa = 3.3NMYY457 pKa = 10.56INKK460 pKa = 7.62THH462 pKa = 6.8WNSTLPSGHH471 pKa = 7.23RR472 pKa = 11.84ATTFINSVLNRR483 pKa = 11.84AYY485 pKa = 10.82LLPFLQVANAFHH497 pKa = 6.54TGDD500 pKa = 4.89DD501 pKa = 3.56VLLCGKK507 pKa = 10.2ADD509 pKa = 3.63YY510 pKa = 7.84ATLINTVPYY519 pKa = 9.77EE520 pKa = 3.98LNKK523 pKa = 9.5TKK525 pKa = 10.72QSFGPSAEE533 pKa = 3.75FLRR536 pKa = 11.84LHH538 pKa = 6.04KK539 pKa = 10.65HH540 pKa = 5.41NDD542 pKa = 3.25QVSGYY547 pKa = 7.28PARR550 pKa = 11.84AISSLVSGNWLSFANPLWQPSLLSIMQQLYY580 pKa = 9.42TISARR585 pKa = 11.84SGLLPYY591 pKa = 10.26IPVTMKK597 pKa = 11.03LEE599 pKa = 3.95VQRR602 pKa = 11.84RR603 pKa = 11.84YY604 pKa = 10.8DD605 pKa = 3.39LRR607 pKa = 11.84SRR609 pKa = 11.84ITNGLFSGDD618 pKa = 3.96IVPSGCPCYY627 pKa = 10.27KK628 pKa = 10.65SNAALLSAVVPDD640 pKa = 4.35TVVKK644 pKa = 10.68APPTFYY650 pKa = 11.06DD651 pKa = 4.84LRR653 pKa = 11.84TLDD656 pKa = 3.59ILKK659 pKa = 8.41QTSPWINSASRR670 pKa = 11.84YY671 pKa = 8.72MDD673 pKa = 3.86LLEE676 pKa = 4.28RR677 pKa = 11.84RR678 pKa = 11.84HH679 pKa = 5.83MEE681 pKa = 3.87SDD683 pKa = 3.3NKK685 pKa = 10.36NVIYY689 pKa = 10.28SIQYY693 pKa = 9.77LPSKK697 pKa = 8.38MLPMIDD703 pKa = 4.33VDD705 pKa = 4.23PADD708 pKa = 3.84ATPLRR713 pKa = 11.84KK714 pKa = 9.04RR715 pKa = 11.84YY716 pKa = 9.37HH717 pKa = 5.94PRR719 pKa = 11.84SHH721 pKa = 7.02IAHH724 pKa = 7.37PLPRR728 pKa = 11.84DD729 pKa = 3.3AHH731 pKa = 5.86LKK733 pKa = 8.18EE734 pKa = 4.81LRR736 pKa = 11.84FATCRR741 pKa = 11.84VGPATAIRR749 pKa = 11.84LGSLWPANRR758 pKa = 11.84INLIKK763 pKa = 10.1PVYY766 pKa = 9.27VV767 pKa = 3.37
Molecular weight: 87.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
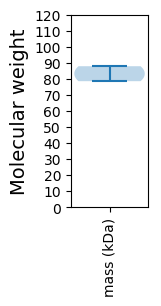
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1476 |
709 |
767 |
738.0 |
83.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.249 ± 0.885 | 1.965 ± 0.199 |
5.691 ± 0.067 | 3.32 ± 0.055 |
4.133 ± 0.381 | 4.607 ± 1.166 |
3.32 ± 0.261 | 6.369 ± 0.016 |
3.862 ± 0.656 | 10.434 ± 1.129 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.168 ± 0.038 | 6.098 ± 0.285 |
6.978 ± 0.465 | 4.472 ± 0.442 |
6.301 ± 0.275 | 7.182 ± 0.403 |
5.691 ± 0.067 | 5.488 ± 0.093 |
1.016 ± 0.227 | 3.659 ± 0.405 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
