
Flavobacteriaceae bacterium
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; unclassified Flavobacteriaceae
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
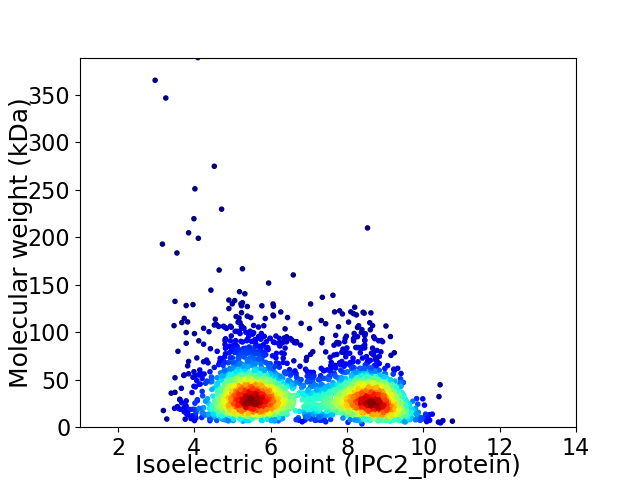
Virtual 2D-PAGE plot for 3010 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A9UP63|A0A3A9UP63_9FLAO Phosphate transport system permease protein OS=Flavobacteriaceae bacterium OX=1871037 GN=pstC PE=3 SV=1
MM1 pKa = 7.31TKK3 pKa = 9.94IIVLFLLVLALKK15 pKa = 8.7ITPSHH20 pKa = 5.97SQTTLTAGDD29 pKa = 3.73IAITGYY35 pKa = 8.31NTDD38 pKa = 4.07GDD40 pKa = 4.11DD41 pKa = 3.3QVAFVLLTDD50 pKa = 3.61ITVGTEE56 pKa = 3.16IRR58 pKa = 11.84FTDD61 pKa = 4.86RR62 pKa = 11.84GWLDD66 pKa = 3.31TNAFRR71 pKa = 11.84IGNTGRR77 pKa = 11.84EE78 pKa = 4.24GTLIWVADD86 pKa = 3.84TDD88 pKa = 4.66LSCGSQIILTSANDD102 pKa = 3.78GTLTISPNIGSLTEE116 pKa = 3.74VDD118 pKa = 3.66DD119 pKa = 4.75FEE121 pKa = 6.1IRR123 pKa = 11.84GQGDD127 pKa = 3.3QILAYY132 pKa = 10.19QGTDD136 pKa = 3.23DD137 pKa = 4.86SPTFIYY143 pKa = 10.68ALNFNNPGWSTTAGNQQEE161 pKa = 4.46SALPIGLTDD170 pKa = 3.98GVNSVDD176 pKa = 3.57ISGDD180 pKa = 3.17IDD182 pKa = 3.54NGTYY186 pKa = 10.38NCAVTTLPDD195 pKa = 4.71AILASVSDD203 pKa = 3.66AANWNTSDD211 pKa = 4.01GDD213 pKa = 4.19GNQSLTLGQCLFSCTSIIQTVLTAGDD239 pKa = 3.78IVITGYY245 pKa = 8.26NTDD248 pKa = 3.98GNDD251 pKa = 2.89QVAFVLLTDD260 pKa = 3.83ITAGTEE266 pKa = 3.43IRR268 pKa = 11.84FTDD271 pKa = 4.45RR272 pKa = 11.84GWLDD276 pKa = 2.98TDD278 pKa = 3.79AFRR281 pKa = 11.84VGNTDD286 pKa = 4.33RR287 pKa = 11.84EE288 pKa = 4.59GTLIWTANTDD298 pKa = 4.22LSCGTQIILTSANNGTLTISPNTGILTEE326 pKa = 4.38EE327 pKa = 4.62DD328 pKa = 3.38DD329 pKa = 4.8FEE331 pKa = 5.37IRR333 pKa = 11.84GQGDD337 pKa = 3.3QILAYY342 pKa = 10.19QGTDD346 pKa = 3.23DD347 pKa = 4.86SPTFIYY353 pKa = 10.68ALNFNNPGWSVTAGNQQEE371 pKa = 4.41SALPIGLADD380 pKa = 3.87GVNSVDD386 pKa = 3.76ISGDD390 pKa = 3.25IDD392 pKa = 3.51NGAYY396 pKa = 10.23DD397 pKa = 4.08CAVTTSPEE405 pKa = 4.51LILTAVSDD413 pKa = 3.61ATNWDD418 pKa = 3.64TSDD421 pKa = 3.34GGGNQSLTLGLCTFDD436 pKa = 5.08CSVICPTTTTWNGTTWDD453 pKa = 3.59NGIPNTTVAAIINGAYY469 pKa = 5.58TTGVNGNISACSLAVNSGFRR489 pKa = 11.84LSISNSTFIEE499 pKa = 4.03IEE501 pKa = 3.75SDD503 pKa = 3.76VVINGEE509 pKa = 4.2IIVEE513 pKa = 4.08SSGNFVQNIDD523 pKa = 3.25SSTYY527 pKa = 8.08TNNGAMSRR535 pKa = 11.84VNKK538 pKa = 7.25VTPVKK543 pKa = 10.46QDD545 pKa = 2.81WFFFTYY551 pKa = 9.48WSSPVSGLTVDD562 pKa = 5.79DD563 pKa = 4.49VFATNPANRR572 pKa = 11.84RR573 pKa = 11.84FIFNANNYY581 pKa = 10.09LDD583 pKa = 4.7LNEE586 pKa = 6.06DD587 pKa = 4.3GFDD590 pKa = 4.09DD591 pKa = 5.09DD592 pKa = 4.92ANAYY596 pKa = 9.16EE597 pKa = 4.79LVSGSDD603 pKa = 3.44PLIPGVGYY611 pKa = 10.55AITEE615 pKa = 4.01NQQFFIPGSTAQATFDD631 pKa = 3.51GTFNNGLIEE640 pKa = 4.14VPIAYY645 pKa = 9.98DD646 pKa = 3.43SANVAHH652 pKa = 6.62YY653 pKa = 10.88NFIGNPYY660 pKa = 9.09PSAIDD665 pKa = 3.56FEE667 pKa = 4.92IFQATNSSLIGGIAYY682 pKa = 9.41LWSQSTPPSANNPGNQTVNFSQNDD706 pKa = 3.63YY707 pKa = 9.45ATYY710 pKa = 9.75TIGSGGAAGASGIIPTQYY728 pKa = 10.02IPSGQGFFIPSVGAGNAVFKK748 pKa = 11.18NSMRR752 pKa = 11.84VASIDD757 pKa = 3.58SNNQFFGTEE766 pKa = 4.17EE767 pKa = 3.78NSLTLNSNPTVNSNDD782 pKa = 3.6LLIDD786 pKa = 3.74NEE788 pKa = 4.09NKK790 pKa = 9.76IWINLKK796 pKa = 10.1SDD798 pKa = 3.16NGIFNQILVAYY809 pKa = 9.98VGGATDD815 pKa = 4.12AYY817 pKa = 11.03DD818 pKa = 3.84GFSYY822 pKa = 10.47DD823 pKa = 3.61APRR826 pKa = 11.84VLPIGTSAILYY837 pKa = 7.44TFIEE841 pKa = 4.82DD842 pKa = 4.87DD843 pKa = 3.52EE844 pKa = 6.89DD845 pKa = 4.91DD846 pKa = 3.68IKK848 pKa = 11.23FVIQGKK854 pKa = 9.89DD855 pKa = 2.89INSINEE861 pKa = 3.63NEE863 pKa = 4.28IIHH866 pKa = 6.7LGFEE870 pKa = 4.52TNIEE874 pKa = 4.19VPTLYY879 pKa = 10.17TLSLDD884 pKa = 3.56QFEE887 pKa = 4.91GAFIEE892 pKa = 4.38NSTIFLKK899 pKa = 11.09DD900 pKa = 3.2NLLDD904 pKa = 3.66VMHH907 pKa = 6.8NLSEE911 pKa = 4.15GDD913 pKa = 3.68YY914 pKa = 10.51EE915 pKa = 4.39FTSEE919 pKa = 3.94VGTFEE924 pKa = 5.22EE925 pKa = 4.67RR926 pKa = 11.84FQIQFVSEE934 pKa = 4.08TLSIDD939 pKa = 3.44EE940 pKa = 4.4NLVIEE945 pKa = 4.36NEE947 pKa = 4.11LVIIEE952 pKa = 4.72LNNNDD957 pKa = 3.14VQFKK961 pKa = 10.71VSGNLEE967 pKa = 3.94MEE969 pKa = 4.48SIKK972 pKa = 10.7IIDD975 pKa = 3.65LNGRR979 pKa = 11.84VLYY982 pKa = 10.36NFKK985 pKa = 10.95AQGSDD990 pKa = 2.77NTYY993 pKa = 11.27NLSKK997 pKa = 10.68LNNSVYY1003 pKa = 10.12IAQIRR1008 pKa = 11.84LTNGVLISKK1017 pKa = 10.2KK1018 pKa = 10.0ALKK1021 pKa = 10.59RR1022 pKa = 11.84NN1023 pKa = 3.56
MM1 pKa = 7.31TKK3 pKa = 9.94IIVLFLLVLALKK15 pKa = 8.7ITPSHH20 pKa = 5.97SQTTLTAGDD29 pKa = 3.73IAITGYY35 pKa = 8.31NTDD38 pKa = 4.07GDD40 pKa = 4.11DD41 pKa = 3.3QVAFVLLTDD50 pKa = 3.61ITVGTEE56 pKa = 3.16IRR58 pKa = 11.84FTDD61 pKa = 4.86RR62 pKa = 11.84GWLDD66 pKa = 3.31TNAFRR71 pKa = 11.84IGNTGRR77 pKa = 11.84EE78 pKa = 4.24GTLIWVADD86 pKa = 3.84TDD88 pKa = 4.66LSCGSQIILTSANDD102 pKa = 3.78GTLTISPNIGSLTEE116 pKa = 3.74VDD118 pKa = 3.66DD119 pKa = 4.75FEE121 pKa = 6.1IRR123 pKa = 11.84GQGDD127 pKa = 3.3QILAYY132 pKa = 10.19QGTDD136 pKa = 3.23DD137 pKa = 4.86SPTFIYY143 pKa = 10.68ALNFNNPGWSTTAGNQQEE161 pKa = 4.46SALPIGLTDD170 pKa = 3.98GVNSVDD176 pKa = 3.57ISGDD180 pKa = 3.17IDD182 pKa = 3.54NGTYY186 pKa = 10.38NCAVTTLPDD195 pKa = 4.71AILASVSDD203 pKa = 3.66AANWNTSDD211 pKa = 4.01GDD213 pKa = 4.19GNQSLTLGQCLFSCTSIIQTVLTAGDD239 pKa = 3.78IVITGYY245 pKa = 8.26NTDD248 pKa = 3.98GNDD251 pKa = 2.89QVAFVLLTDD260 pKa = 3.83ITAGTEE266 pKa = 3.43IRR268 pKa = 11.84FTDD271 pKa = 4.45RR272 pKa = 11.84GWLDD276 pKa = 2.98TDD278 pKa = 3.79AFRR281 pKa = 11.84VGNTDD286 pKa = 4.33RR287 pKa = 11.84EE288 pKa = 4.59GTLIWTANTDD298 pKa = 4.22LSCGTQIILTSANNGTLTISPNTGILTEE326 pKa = 4.38EE327 pKa = 4.62DD328 pKa = 3.38DD329 pKa = 4.8FEE331 pKa = 5.37IRR333 pKa = 11.84GQGDD337 pKa = 3.3QILAYY342 pKa = 10.19QGTDD346 pKa = 3.23DD347 pKa = 4.86SPTFIYY353 pKa = 10.68ALNFNNPGWSVTAGNQQEE371 pKa = 4.41SALPIGLADD380 pKa = 3.87GVNSVDD386 pKa = 3.76ISGDD390 pKa = 3.25IDD392 pKa = 3.51NGAYY396 pKa = 10.23DD397 pKa = 4.08CAVTTSPEE405 pKa = 4.51LILTAVSDD413 pKa = 3.61ATNWDD418 pKa = 3.64TSDD421 pKa = 3.34GGGNQSLTLGLCTFDD436 pKa = 5.08CSVICPTTTTWNGTTWDD453 pKa = 3.59NGIPNTTVAAIINGAYY469 pKa = 5.58TTGVNGNISACSLAVNSGFRR489 pKa = 11.84LSISNSTFIEE499 pKa = 4.03IEE501 pKa = 3.75SDD503 pKa = 3.76VVINGEE509 pKa = 4.2IIVEE513 pKa = 4.08SSGNFVQNIDD523 pKa = 3.25SSTYY527 pKa = 8.08TNNGAMSRR535 pKa = 11.84VNKK538 pKa = 7.25VTPVKK543 pKa = 10.46QDD545 pKa = 2.81WFFFTYY551 pKa = 9.48WSSPVSGLTVDD562 pKa = 5.79DD563 pKa = 4.49VFATNPANRR572 pKa = 11.84RR573 pKa = 11.84FIFNANNYY581 pKa = 10.09LDD583 pKa = 4.7LNEE586 pKa = 6.06DD587 pKa = 4.3GFDD590 pKa = 4.09DD591 pKa = 5.09DD592 pKa = 4.92ANAYY596 pKa = 9.16EE597 pKa = 4.79LVSGSDD603 pKa = 3.44PLIPGVGYY611 pKa = 10.55AITEE615 pKa = 4.01NQQFFIPGSTAQATFDD631 pKa = 3.51GTFNNGLIEE640 pKa = 4.14VPIAYY645 pKa = 9.98DD646 pKa = 3.43SANVAHH652 pKa = 6.62YY653 pKa = 10.88NFIGNPYY660 pKa = 9.09PSAIDD665 pKa = 3.56FEE667 pKa = 4.92IFQATNSSLIGGIAYY682 pKa = 9.41LWSQSTPPSANNPGNQTVNFSQNDD706 pKa = 3.63YY707 pKa = 9.45ATYY710 pKa = 9.75TIGSGGAAGASGIIPTQYY728 pKa = 10.02IPSGQGFFIPSVGAGNAVFKK748 pKa = 11.18NSMRR752 pKa = 11.84VASIDD757 pKa = 3.58SNNQFFGTEE766 pKa = 4.17EE767 pKa = 3.78NSLTLNSNPTVNSNDD782 pKa = 3.6LLIDD786 pKa = 3.74NEE788 pKa = 4.09NKK790 pKa = 9.76IWINLKK796 pKa = 10.1SDD798 pKa = 3.16NGIFNQILVAYY809 pKa = 9.98VGGATDD815 pKa = 4.12AYY817 pKa = 11.03DD818 pKa = 3.84GFSYY822 pKa = 10.47DD823 pKa = 3.61APRR826 pKa = 11.84VLPIGTSAILYY837 pKa = 7.44TFIEE841 pKa = 4.82DD842 pKa = 4.87DD843 pKa = 3.52EE844 pKa = 6.89DD845 pKa = 4.91DD846 pKa = 3.68IKK848 pKa = 11.23FVIQGKK854 pKa = 9.89DD855 pKa = 2.89INSINEE861 pKa = 3.63NEE863 pKa = 4.28IIHH866 pKa = 6.7LGFEE870 pKa = 4.52TNIEE874 pKa = 4.19VPTLYY879 pKa = 10.17TLSLDD884 pKa = 3.56QFEE887 pKa = 4.91GAFIEE892 pKa = 4.38NSTIFLKK899 pKa = 11.09DD900 pKa = 3.2NLLDD904 pKa = 3.66VMHH907 pKa = 6.8NLSEE911 pKa = 4.15GDD913 pKa = 3.68YY914 pKa = 10.51EE915 pKa = 4.39FTSEE919 pKa = 3.94VGTFEE924 pKa = 5.22EE925 pKa = 4.67RR926 pKa = 11.84FQIQFVSEE934 pKa = 4.08TLSIDD939 pKa = 3.44EE940 pKa = 4.4NLVIEE945 pKa = 4.36NEE947 pKa = 4.11LVIIEE952 pKa = 4.72LNNNDD957 pKa = 3.14VQFKK961 pKa = 10.71VSGNLEE967 pKa = 3.94MEE969 pKa = 4.48SIKK972 pKa = 10.7IIDD975 pKa = 3.65LNGRR979 pKa = 11.84VLYY982 pKa = 10.36NFKK985 pKa = 10.95AQGSDD990 pKa = 2.77NTYY993 pKa = 11.27NLSKK997 pKa = 10.68LNNSVYY1003 pKa = 10.12IAQIRR1008 pKa = 11.84LTNGVLISKK1017 pKa = 10.2KK1018 pKa = 10.0ALKK1021 pKa = 10.59RR1022 pKa = 11.84NN1023 pKa = 3.56
Molecular weight: 110.12 kDa
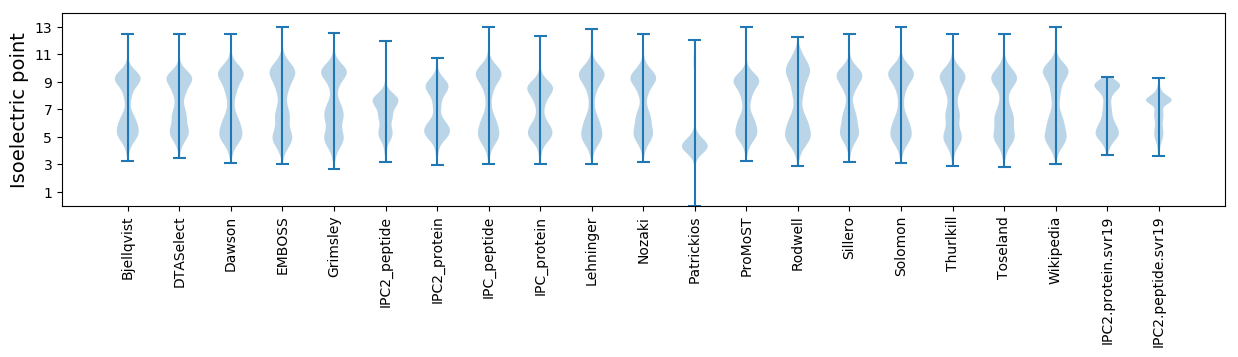
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A9UMP3|A0A3A9UMP3_9FLAO RDD family protein OS=Flavobacteriaceae bacterium OX=1871037 GN=D1817_14665 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.92GRR40 pKa = 11.84KK41 pKa = 8.34KK42 pKa = 10.22ISVSSEE48 pKa = 3.61LRR50 pKa = 11.84HH51 pKa = 6.16KK52 pKa = 10.53KK53 pKa = 10.04
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.92GRR40 pKa = 11.84KK41 pKa = 8.34KK42 pKa = 10.22ISVSSEE48 pKa = 3.61LRR50 pKa = 11.84HH51 pKa = 6.16KK52 pKa = 10.53KK53 pKa = 10.04
Molecular weight: 6.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1019847 |
29 |
3644 |
338.8 |
38.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.87 ± 0.043 | 0.69 ± 0.015 |
5.505 ± 0.044 | 6.423 ± 0.048 |
5.348 ± 0.044 | 6.183 ± 0.051 |
1.715 ± 0.023 | 8.892 ± 0.049 |
7.943 ± 0.079 | 9.368 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.945 ± 0.024 | 6.775 ± 0.057 |
3.245 ± 0.025 | 3.308 ± 0.026 |
3.447 ± 0.028 | 6.6 ± 0.039 |
5.905 ± 0.064 | 5.82 ± 0.033 |
1.001 ± 0.018 | 4.018 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
