
Cycloclasticus sp. (strain P1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Piscirickettsiaceae; Cycloclasticus; unclassified Cycloclasticus
Average proteome isoelectric point is 6.21
Get precalculated fractions of proteins
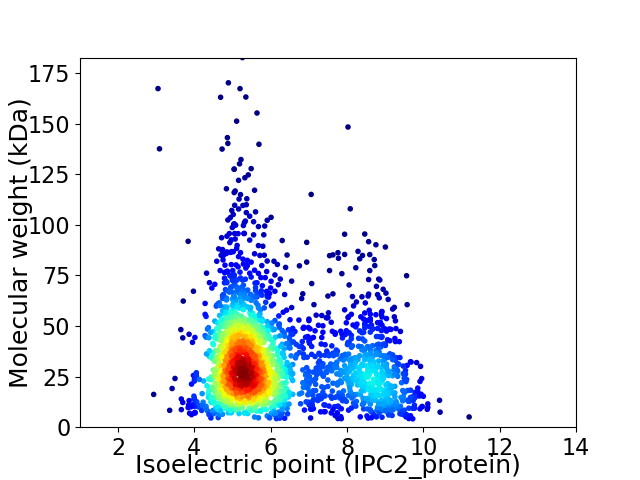
Virtual 2D-PAGE plot for 2247 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0C0P0|K0C0P0_CYCSP UvrABC system protein B OS=Cycloclasticus sp. (strain P1) OX=385025 GN=uvrB PE=3 SV=1
MM1 pKa = 7.75KK2 pKa = 10.16LNKK5 pKa = 9.48IQPSLNNQKK14 pKa = 10.15GVATLLVSIILLIGVTLITVFAARR38 pKa = 11.84VGVMDD43 pKa = 3.78QRR45 pKa = 11.84ISANEE50 pKa = 3.76YY51 pKa = 8.71RR52 pKa = 11.84HH53 pKa = 6.5KK54 pKa = 10.28EE55 pKa = 3.79AQAAADD61 pKa = 3.58AALEE65 pKa = 3.86QAAAFINEE73 pKa = 3.95NTGLYY78 pKa = 10.32DD79 pKa = 3.39GTVGGTYY86 pKa = 9.9PWVEE90 pKa = 4.02CVGGYY95 pKa = 8.65TSLFPCTNGSDD106 pKa = 4.17TYY108 pKa = 11.81NLAYY112 pKa = 10.71DD113 pKa = 4.0NDD115 pKa = 4.28LTTLTTIEE123 pKa = 4.3PLQLGATPTAVDD135 pKa = 4.28LSSGVVSNTYY145 pKa = 8.48LTYY148 pKa = 9.51TNSGNVMTAIGTGEE162 pKa = 4.12SLDD165 pKa = 3.92GTGNAYY171 pKa = 10.25AQISYY176 pKa = 8.06TQITLLTSEE185 pKa = 4.6EE186 pKa = 4.44VPPIMTPIIEE196 pKa = 4.45LGGTFTIVADD206 pKa = 4.06PNGGGKK212 pKa = 9.75GVPVSAWVATFDD224 pKa = 3.55TSGGLGSWQTCQLDD238 pKa = 3.57AFLDD242 pKa = 3.83NSGDD246 pKa = 3.57ICSEE250 pKa = 4.07TLDD253 pKa = 4.93DD254 pKa = 3.6TDD256 pKa = 2.8SWKK259 pKa = 10.84FCDD262 pKa = 4.5CDD264 pKa = 3.76GDD266 pKa = 4.15KK267 pKa = 10.99LSEE270 pKa = 4.29PGKK273 pKa = 9.88VNNDD277 pKa = 2.45IVQADD282 pKa = 3.99PGDD285 pKa = 4.15FPASVLSYY293 pKa = 10.04LFPNFDD299 pKa = 3.36SFQDD303 pKa = 3.6VVNIADD309 pKa = 3.97VEE311 pKa = 4.29LTNCSGLQALASGWSTSKK329 pKa = 10.54IVTVDD334 pKa = 3.88GDD336 pKa = 3.87CTMPSNSIIGSRR348 pKa = 11.84DD349 pKa = 3.0GPLILVVSGHH359 pKa = 6.45LKK361 pKa = 10.17INANSDD367 pKa = 3.63FYY369 pKa = 11.49GIALGLSDD377 pKa = 3.62VTLNGGVKK385 pKa = 9.15VHH387 pKa = 6.63GSLLAEE393 pKa = 5.16DD394 pKa = 3.99STKK397 pKa = 10.31LTTGGYY403 pKa = 8.81AQVYY407 pKa = 10.29DD408 pKa = 4.17EE409 pKa = 5.31FVLGSIVEE417 pKa = 4.23DD418 pKa = 4.09LSNLGLAKK426 pKa = 10.03QKK428 pKa = 10.95YY429 pKa = 8.45SWTDD433 pKa = 2.98IQPP436 pKa = 3.25
MM1 pKa = 7.75KK2 pKa = 10.16LNKK5 pKa = 9.48IQPSLNNQKK14 pKa = 10.15GVATLLVSIILLIGVTLITVFAARR38 pKa = 11.84VGVMDD43 pKa = 3.78QRR45 pKa = 11.84ISANEE50 pKa = 3.76YY51 pKa = 8.71RR52 pKa = 11.84HH53 pKa = 6.5KK54 pKa = 10.28EE55 pKa = 3.79AQAAADD61 pKa = 3.58AALEE65 pKa = 3.86QAAAFINEE73 pKa = 3.95NTGLYY78 pKa = 10.32DD79 pKa = 3.39GTVGGTYY86 pKa = 9.9PWVEE90 pKa = 4.02CVGGYY95 pKa = 8.65TSLFPCTNGSDD106 pKa = 4.17TYY108 pKa = 11.81NLAYY112 pKa = 10.71DD113 pKa = 4.0NDD115 pKa = 4.28LTTLTTIEE123 pKa = 4.3PLQLGATPTAVDD135 pKa = 4.28LSSGVVSNTYY145 pKa = 8.48LTYY148 pKa = 9.51TNSGNVMTAIGTGEE162 pKa = 4.12SLDD165 pKa = 3.92GTGNAYY171 pKa = 10.25AQISYY176 pKa = 8.06TQITLLTSEE185 pKa = 4.6EE186 pKa = 4.44VPPIMTPIIEE196 pKa = 4.45LGGTFTIVADD206 pKa = 4.06PNGGGKK212 pKa = 9.75GVPVSAWVATFDD224 pKa = 3.55TSGGLGSWQTCQLDD238 pKa = 3.57AFLDD242 pKa = 3.83NSGDD246 pKa = 3.57ICSEE250 pKa = 4.07TLDD253 pKa = 4.93DD254 pKa = 3.6TDD256 pKa = 2.8SWKK259 pKa = 10.84FCDD262 pKa = 4.5CDD264 pKa = 3.76GDD266 pKa = 4.15KK267 pKa = 10.99LSEE270 pKa = 4.29PGKK273 pKa = 9.88VNNDD277 pKa = 2.45IVQADD282 pKa = 3.99PGDD285 pKa = 4.15FPASVLSYY293 pKa = 10.04LFPNFDD299 pKa = 3.36SFQDD303 pKa = 3.6VVNIADD309 pKa = 3.97VEE311 pKa = 4.29LTNCSGLQALASGWSTSKK329 pKa = 10.54IVTVDD334 pKa = 3.88GDD336 pKa = 3.87CTMPSNSIIGSRR348 pKa = 11.84DD349 pKa = 3.0GPLILVVSGHH359 pKa = 6.45LKK361 pKa = 10.17INANSDD367 pKa = 3.63FYY369 pKa = 11.49GIALGLSDD377 pKa = 3.62VTLNGGVKK385 pKa = 9.15VHH387 pKa = 6.63GSLLAEE393 pKa = 5.16DD394 pKa = 3.99STKK397 pKa = 10.31LTTGGYY403 pKa = 8.81AQVYY407 pKa = 10.29DD408 pKa = 4.17EE409 pKa = 5.31FVLGSIVEE417 pKa = 4.23DD418 pKa = 4.09LSNLGLAKK426 pKa = 10.03QKK428 pKa = 10.95YY429 pKa = 8.45SWTDD433 pKa = 2.98IQPP436 pKa = 3.25
Molecular weight: 45.91 kDa
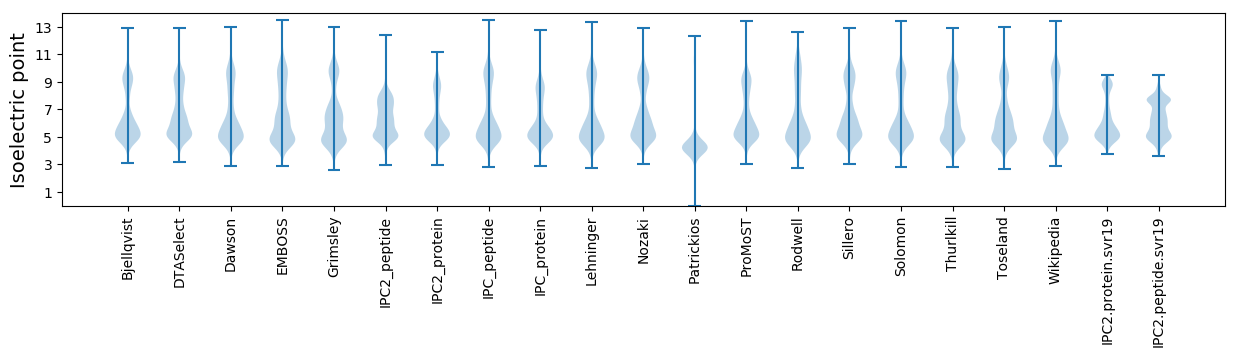
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0C4V4|K0C4V4_CYCSP Replicative DNA helicase OS=Cycloclasticus sp. (strain P1) OX=385025 GN=dnaB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27SGRR28 pKa = 11.84KK29 pKa = 8.85IINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27SGRR28 pKa = 11.84KK29 pKa = 8.85IINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
715765 |
38 |
1645 |
318.5 |
35.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.419 ± 0.049 | 1.017 ± 0.017 |
5.647 ± 0.045 | 6.291 ± 0.057 |
4.056 ± 0.036 | 6.917 ± 0.049 |
2.262 ± 0.028 | 6.747 ± 0.043 |
5.777 ± 0.048 | 10.491 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.576 ± 0.024 | 4.291 ± 0.031 |
3.93 ± 0.031 | 4.22 ± 0.042 |
4.438 ± 0.041 | 6.528 ± 0.044 |
5.418 ± 0.041 | 6.959 ± 0.048 |
1.144 ± 0.02 | 2.87 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
