
Phyllobacterium sp. YR531
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Phyllobacterium; unclassified Phyllobacterium
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
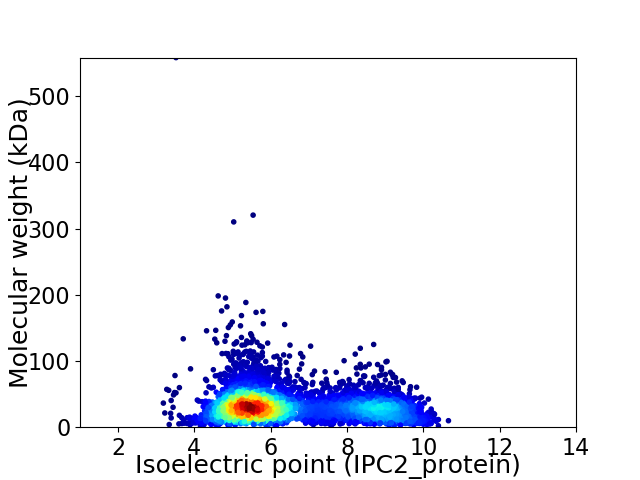
Virtual 2D-PAGE plot for 4844 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J3C622|J3C622_9RHIZ Peptidase T OS=Phyllobacterium sp. YR531 OX=1144343 GN=PMI41_04132 PE=3 SV=1
MM1 pKa = 7.28VLTDD5 pKa = 4.18GDD7 pKa = 3.97DD8 pKa = 3.85VYY10 pKa = 11.39TGTAGNDD17 pKa = 3.04LVYY20 pKa = 11.05ALNGNDD26 pKa = 3.71KK27 pKa = 11.0VSGGLGNDD35 pKa = 3.17QFYY38 pKa = 11.19GGAGSDD44 pKa = 3.53ILDD47 pKa = 4.36GGDD50 pKa = 4.16GDD52 pKa = 4.77DD53 pKa = 5.33LIYY56 pKa = 10.53GYY58 pKa = 10.73GSSGTDD64 pKa = 3.12GGPDD68 pKa = 3.28VPGNDD73 pKa = 3.36TLLGGRR79 pKa = 11.84GEE81 pKa = 4.23DD82 pKa = 3.33QLYY85 pKa = 11.04SGIGNDD91 pKa = 4.19FLDD94 pKa = 4.62GGADD98 pKa = 3.67DD99 pKa = 6.04DD100 pKa = 5.25YY101 pKa = 11.56ISSGDD106 pKa = 3.54GNDD109 pKa = 3.4RR110 pKa = 11.84LFGSDD115 pKa = 5.47GDD117 pKa = 3.94DD118 pKa = 3.59TLNGGAGNDD127 pKa = 3.43ILNGGTGEE135 pKa = 4.14DD136 pKa = 4.09RR137 pKa = 11.84MTGGTGSDD145 pKa = 2.75IYY147 pKa = 11.44YY148 pKa = 10.29VDD150 pKa = 3.83SADD153 pKa = 4.32DD154 pKa = 3.72EE155 pKa = 4.91VNEE158 pKa = 4.25FSGGGIDD165 pKa = 4.71RR166 pKa = 11.84VISSISFSLTSDD178 pKa = 3.52SFVVSGDD185 pKa = 3.48VEE187 pKa = 4.45YY188 pKa = 9.64LTLSGSANIDD198 pKa = 3.36GQGNSLNNLIYY209 pKa = 10.85GNNGNNALRR218 pKa = 11.84GWGGHH223 pKa = 4.46DD224 pKa = 4.65TIYY227 pKa = 10.87GWAGNDD233 pKa = 3.09ILDD236 pKa = 4.03GGFGKK241 pKa = 10.49DD242 pKa = 3.11MLVGGEE248 pKa = 4.41GNDD251 pKa = 3.36TYY253 pKa = 11.6YY254 pKa = 11.08VDD256 pKa = 3.57SAGDD260 pKa = 3.4RR261 pKa = 11.84VVEE264 pKa = 4.06ARR266 pKa = 11.84VTGIDD271 pKa = 3.35TVIASVSFSTVGQYY285 pKa = 9.83IEE287 pKa = 4.99NVTLTGSANINATGNSLGNKK307 pKa = 9.22LVGNSGNNSLDD318 pKa = 3.25GKK320 pKa = 9.1TGADD324 pKa = 3.54SMSGGAGSDD333 pKa = 2.97KK334 pKa = 11.12YY335 pKa = 11.47YY336 pKa = 10.32IDD338 pKa = 3.58NAGDD342 pKa = 3.85KK343 pKa = 10.73VIEE346 pKa = 4.38GNVAGTDD353 pKa = 3.68TVSSSVSFATGGQHH367 pKa = 6.58IEE369 pKa = 4.15NVLLTGSASINATGNALNNYY389 pKa = 9.48LVGNSGANIISGGAGNDD406 pKa = 3.72LLTGGAGRR414 pKa = 11.84DD415 pKa = 2.86AFVFNTAPASNNRR428 pKa = 11.84DD429 pKa = 3.48TITDD433 pKa = 3.97FNVVADD439 pKa = 5.04TIWMDD444 pKa = 3.35NAIFTKK450 pKa = 10.71LGATGALAATAFHH463 pKa = 7.12IGTAAADD470 pKa = 3.38ASDD473 pKa = 4.11RR474 pKa = 11.84IIYY477 pKa = 10.59NSTTGALIYY486 pKa = 10.36DD487 pKa = 4.31SNGTAAGGATQFATLSKK504 pKa = 10.85GLALTNADD512 pKa = 3.77FLVII516 pKa = 4.33
MM1 pKa = 7.28VLTDD5 pKa = 4.18GDD7 pKa = 3.97DD8 pKa = 3.85VYY10 pKa = 11.39TGTAGNDD17 pKa = 3.04LVYY20 pKa = 11.05ALNGNDD26 pKa = 3.71KK27 pKa = 11.0VSGGLGNDD35 pKa = 3.17QFYY38 pKa = 11.19GGAGSDD44 pKa = 3.53ILDD47 pKa = 4.36GGDD50 pKa = 4.16GDD52 pKa = 4.77DD53 pKa = 5.33LIYY56 pKa = 10.53GYY58 pKa = 10.73GSSGTDD64 pKa = 3.12GGPDD68 pKa = 3.28VPGNDD73 pKa = 3.36TLLGGRR79 pKa = 11.84GEE81 pKa = 4.23DD82 pKa = 3.33QLYY85 pKa = 11.04SGIGNDD91 pKa = 4.19FLDD94 pKa = 4.62GGADD98 pKa = 3.67DD99 pKa = 6.04DD100 pKa = 5.25YY101 pKa = 11.56ISSGDD106 pKa = 3.54GNDD109 pKa = 3.4RR110 pKa = 11.84LFGSDD115 pKa = 5.47GDD117 pKa = 3.94DD118 pKa = 3.59TLNGGAGNDD127 pKa = 3.43ILNGGTGEE135 pKa = 4.14DD136 pKa = 4.09RR137 pKa = 11.84MTGGTGSDD145 pKa = 2.75IYY147 pKa = 11.44YY148 pKa = 10.29VDD150 pKa = 3.83SADD153 pKa = 4.32DD154 pKa = 3.72EE155 pKa = 4.91VNEE158 pKa = 4.25FSGGGIDD165 pKa = 4.71RR166 pKa = 11.84VISSISFSLTSDD178 pKa = 3.52SFVVSGDD185 pKa = 3.48VEE187 pKa = 4.45YY188 pKa = 9.64LTLSGSANIDD198 pKa = 3.36GQGNSLNNLIYY209 pKa = 10.85GNNGNNALRR218 pKa = 11.84GWGGHH223 pKa = 4.46DD224 pKa = 4.65TIYY227 pKa = 10.87GWAGNDD233 pKa = 3.09ILDD236 pKa = 4.03GGFGKK241 pKa = 10.49DD242 pKa = 3.11MLVGGEE248 pKa = 4.41GNDD251 pKa = 3.36TYY253 pKa = 11.6YY254 pKa = 11.08VDD256 pKa = 3.57SAGDD260 pKa = 3.4RR261 pKa = 11.84VVEE264 pKa = 4.06ARR266 pKa = 11.84VTGIDD271 pKa = 3.35TVIASVSFSTVGQYY285 pKa = 9.83IEE287 pKa = 4.99NVTLTGSANINATGNSLGNKK307 pKa = 9.22LVGNSGNNSLDD318 pKa = 3.25GKK320 pKa = 9.1TGADD324 pKa = 3.54SMSGGAGSDD333 pKa = 2.97KK334 pKa = 11.12YY335 pKa = 11.47YY336 pKa = 10.32IDD338 pKa = 3.58NAGDD342 pKa = 3.85KK343 pKa = 10.73VIEE346 pKa = 4.38GNVAGTDD353 pKa = 3.68TVSSSVSFATGGQHH367 pKa = 6.58IEE369 pKa = 4.15NVLLTGSASINATGNALNNYY389 pKa = 9.48LVGNSGANIISGGAGNDD406 pKa = 3.72LLTGGAGRR414 pKa = 11.84DD415 pKa = 2.86AFVFNTAPASNNRR428 pKa = 11.84DD429 pKa = 3.48TITDD433 pKa = 3.97FNVVADD439 pKa = 5.04TIWMDD444 pKa = 3.35NAIFTKK450 pKa = 10.71LGATGALAATAFHH463 pKa = 7.12IGTAAADD470 pKa = 3.38ASDD473 pKa = 4.11RR474 pKa = 11.84IIYY477 pKa = 10.59NSTTGALIYY486 pKa = 10.36DD487 pKa = 4.31SNGTAAGGATQFATLSKK504 pKa = 10.85GLALTNADD512 pKa = 3.77FLVII516 pKa = 4.33
Molecular weight: 51.89 kDa
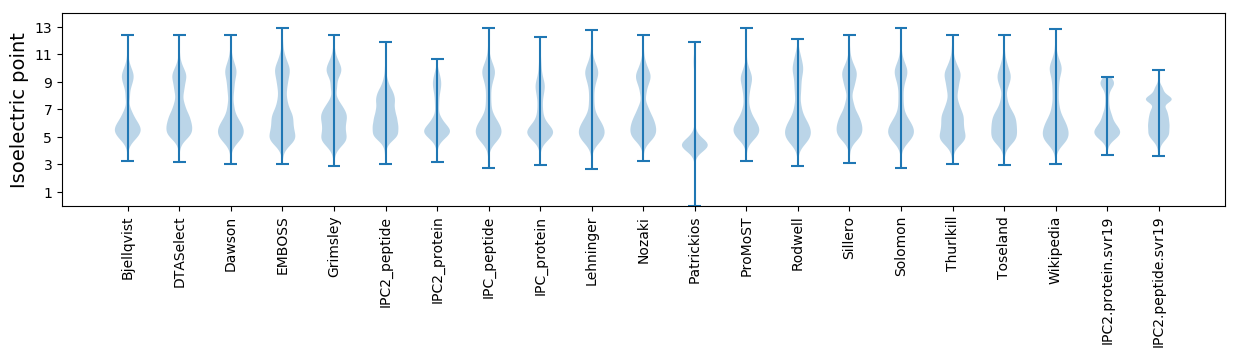
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J2VIT4|J2VIT4_9RHIZ Uncharacterized protein OS=Phyllobacterium sp. YR531 OX=1144343 GN=PMI41_03803 PE=4 SV=1
MM1 pKa = 7.46PRR3 pKa = 11.84SSLKK7 pKa = 10.99YY8 pKa = 8.22IDD10 pKa = 3.41VRR12 pKa = 11.84PRR14 pKa = 11.84PGHH17 pKa = 6.16KK18 pKa = 10.23SQGQLIAGGIVLRR31 pKa = 11.84CALGKK36 pKa = 10.77GGIISRR42 pKa = 11.84KK43 pKa = 9.26RR44 pKa = 11.84EE45 pKa = 3.97GDD47 pKa = 2.93GGTPLASMRR56 pKa = 11.84LLYY59 pKa = 10.59GFKK62 pKa = 10.62RR63 pKa = 11.84PDD65 pKa = 3.15KK66 pKa = 10.11QVRR69 pKa = 11.84ATRR72 pKa = 11.84LLRR75 pKa = 11.84LNPLKK80 pKa = 10.31TNDD83 pKa = 3.15GWCEE87 pKa = 3.64VSNDD91 pKa = 3.22RR92 pKa = 11.84NYY94 pKa = 10.71NRR96 pKa = 11.84QVQIPYY102 pKa = 9.22NASHH106 pKa = 5.52EE107 pKa = 4.57TMWRR111 pKa = 11.84KK112 pKa = 10.26DD113 pKa = 3.77DD114 pKa = 5.27LYY116 pKa = 10.93DD117 pKa = 3.36ICIVLDD123 pKa = 3.34WNIRR127 pKa = 11.84PRR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84AGGSAIFFHH140 pKa = 6.99LARR143 pKa = 11.84PQYY146 pKa = 10.04TPTEE150 pKa = 3.87GCIALNRR157 pKa = 11.84ADD159 pKa = 4.07MNRR162 pKa = 11.84LLPHH166 pKa = 7.03LSDD169 pKa = 3.17KK170 pKa = 10.76TVIRR174 pKa = 11.84VLRR177 pKa = 4.06
MM1 pKa = 7.46PRR3 pKa = 11.84SSLKK7 pKa = 10.99YY8 pKa = 8.22IDD10 pKa = 3.41VRR12 pKa = 11.84PRR14 pKa = 11.84PGHH17 pKa = 6.16KK18 pKa = 10.23SQGQLIAGGIVLRR31 pKa = 11.84CALGKK36 pKa = 10.77GGIISRR42 pKa = 11.84KK43 pKa = 9.26RR44 pKa = 11.84EE45 pKa = 3.97GDD47 pKa = 2.93GGTPLASMRR56 pKa = 11.84LLYY59 pKa = 10.59GFKK62 pKa = 10.62RR63 pKa = 11.84PDD65 pKa = 3.15KK66 pKa = 10.11QVRR69 pKa = 11.84ATRR72 pKa = 11.84LLRR75 pKa = 11.84LNPLKK80 pKa = 10.31TNDD83 pKa = 3.15GWCEE87 pKa = 3.64VSNDD91 pKa = 3.22RR92 pKa = 11.84NYY94 pKa = 10.71NRR96 pKa = 11.84QVQIPYY102 pKa = 9.22NASHH106 pKa = 5.52EE107 pKa = 4.57TMWRR111 pKa = 11.84KK112 pKa = 10.26DD113 pKa = 3.77DD114 pKa = 5.27LYY116 pKa = 10.93DD117 pKa = 3.36ICIVLDD123 pKa = 3.34WNIRR127 pKa = 11.84PRR129 pKa = 11.84RR130 pKa = 11.84RR131 pKa = 11.84AGGSAIFFHH140 pKa = 6.99LARR143 pKa = 11.84PQYY146 pKa = 10.04TPTEE150 pKa = 3.87GCIALNRR157 pKa = 11.84ADD159 pKa = 4.07MNRR162 pKa = 11.84LLPHH166 pKa = 7.03LSDD169 pKa = 3.17KK170 pKa = 10.76TVIRR174 pKa = 11.84VLRR177 pKa = 4.06
Molecular weight: 20.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1500639 |
18 |
5517 |
309.8 |
33.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.004 ± 0.044 | 0.733 ± 0.01 |
5.657 ± 0.03 | 5.682 ± 0.037 |
4.002 ± 0.024 | 8.188 ± 0.04 |
1.972 ± 0.019 | 6.231 ± 0.027 |
4.221 ± 0.032 | 9.875 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.459 ± 0.016 | 3.346 ± 0.031 |
4.708 ± 0.024 | 3.252 ± 0.022 |
6.062 ± 0.039 | 6.128 ± 0.028 |
5.562 ± 0.047 | 7.199 ± 0.03 |
1.278 ± 0.015 | 2.44 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
