
Thermoanaerobaculum aquaticum
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Thermoanaerobaculia; Thermoanaerobaculales; Thermoanaerobaculaceae; Thermoanaerobaculum
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
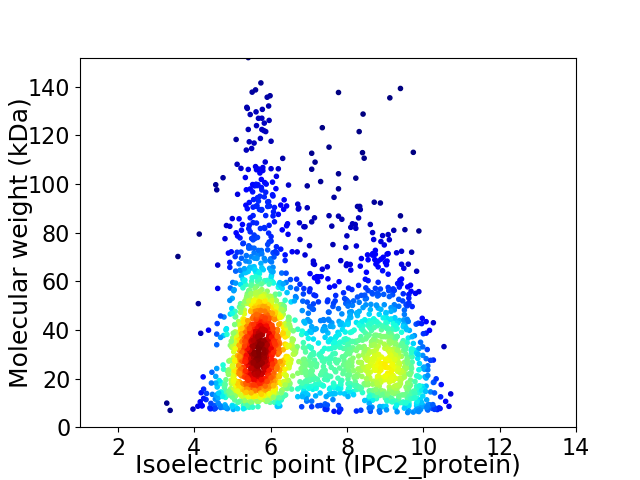
Virtual 2D-PAGE plot for 2252 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
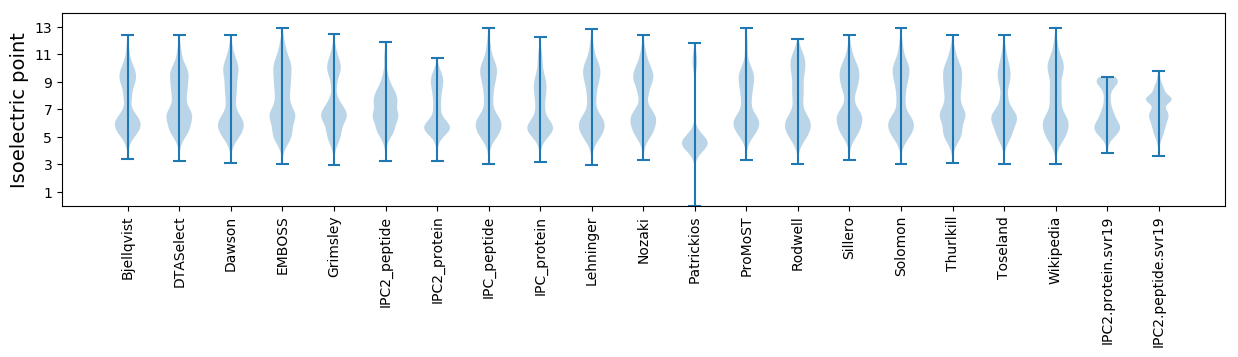
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A062XM57|A0A062XM57_9BACT ABC transmembrane type-1 domain-containing protein OS=Thermoanaerobaculum aquaticum OX=1312852 GN=EG19_05510 PE=3 SV=1
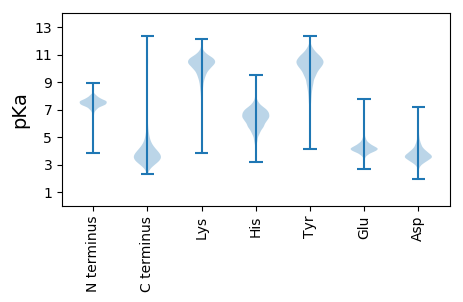
MM1 pKa = 7.76ADD3 pKa = 3.42KK4 pKa = 10.55TNKK7 pKa = 9.53VPLNVPGKK15 pKa = 8.57WYY17 pKa = 10.97VDD19 pKa = 3.75TSCIDD24 pKa = 3.49CDD26 pKa = 3.95VCRR29 pKa = 11.84TTAPNNFKK37 pKa = 11.01ANEE40 pKa = 4.08DD41 pKa = 3.25EE42 pKa = 4.89GYY44 pKa = 10.57SYY46 pKa = 11.38VYY48 pKa = 9.78KK49 pKa = 10.63QPEE52 pKa = 4.27TPEE55 pKa = 4.61EE56 pKa = 3.8EE57 pKa = 4.51AQCQEE62 pKa = 4.42AKK64 pKa = 10.52ASCPVEE70 pKa = 5.03AIGDD74 pKa = 4.17DD75 pKa = 3.79GDD77 pKa = 3.75EE78 pKa = 4.08
MM1 pKa = 7.76ADD3 pKa = 3.42KK4 pKa = 10.55TNKK7 pKa = 9.53VPLNVPGKK15 pKa = 8.57WYY17 pKa = 10.97VDD19 pKa = 3.75TSCIDD24 pKa = 3.49CDD26 pKa = 3.95VCRR29 pKa = 11.84TTAPNNFKK37 pKa = 11.01ANEE40 pKa = 4.08DD41 pKa = 3.25EE42 pKa = 4.89GYY44 pKa = 10.57SYY46 pKa = 11.38VYY48 pKa = 9.78KK49 pKa = 10.63QPEE52 pKa = 4.27TPEE55 pKa = 4.61EE56 pKa = 3.8EE57 pKa = 4.51AQCQEE62 pKa = 4.42AKK64 pKa = 10.52ASCPVEE70 pKa = 5.03AIGDD74 pKa = 4.17DD75 pKa = 3.79GDD77 pKa = 3.75EE78 pKa = 4.08
Molecular weight: 8.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A062XXP5|A0A062XXP5_9BACT Uncharacterized protein OS=Thermoanaerobaculum aquaticum OX=1312852 GN=EG19_09350 PE=4 SV=1
MM1 pKa = 7.5SFRR4 pKa = 11.84HH5 pKa = 5.57AFSLAVLPGLALVLASFAGWVPAARR30 pKa = 11.84DD31 pKa = 3.48NPTYY35 pKa = 10.19FVPLRR40 pKa = 11.84AQLARR45 pKa = 11.84VFTGEE50 pKa = 3.76ASPWLNPQVGCGEE63 pKa = 4.18PFFANPQSALLYY75 pKa = 8.84PPAWGALLLPPEE87 pKa = 3.99QAIGVEE93 pKa = 4.37VGLHH97 pKa = 6.2LMLLALGVARR107 pKa = 11.84LARR110 pKa = 11.84RR111 pKa = 11.84LGGTPTGSLAAAWGAALSGPVLSSAGMLNNLEE143 pKa = 4.06TAAWLPWVWDD153 pKa = 3.56AALSGRR159 pKa = 11.84FGVLALTVTGSFLAAEE175 pKa = 4.43PVLALLGAAGAVWLLPRR192 pKa = 11.84WQAVRR197 pKa = 11.84AVLLGFALCSVQALPMAFWIAGGDD221 pKa = 3.6RR222 pKa = 11.84GPDD225 pKa = 3.21KK226 pKa = 10.74PLEE229 pKa = 4.25AVSLGGVSLGEE240 pKa = 4.06LPALVVPGFPLPPVEE255 pKa = 5.03VRR257 pKa = 11.84FLPIIALPLWLFLALGSLRR276 pKa = 11.84RR277 pKa = 11.84GEE279 pKa = 4.14TARR282 pKa = 11.84TRR284 pKa = 11.84LASLAAVFTFLAVLPALPWGDD305 pKa = 4.04ALWAGLSFGLVRR317 pKa = 11.84LPGRR321 pKa = 11.84FLIPATVALAAVAGSGKK338 pKa = 10.17LPQSRR343 pKa = 11.84KK344 pKa = 7.14WVATALGLGAVGAVVSRR361 pKa = 11.84EE362 pKa = 3.59PWLAAGQGVLAALAPWGMGWAAAGSLFLATYY393 pKa = 7.4TVPVLQLQKK402 pKa = 10.15WKK404 pKa = 9.96PEE406 pKa = 3.8PVLCLSAQSAGRR418 pKa = 11.84LYY420 pKa = 9.77PLPVDD425 pKa = 4.43GVQLRR430 pKa = 11.84WTLEE434 pKa = 3.61RR435 pKa = 11.84GTRR438 pKa = 11.84GAASLGWGYY447 pKa = 11.33SVLLDD452 pKa = 3.53GRR454 pKa = 11.84SLARR458 pKa = 11.84SFAPVTNRR466 pKa = 11.84ALAQHH471 pKa = 7.25LAQADD476 pKa = 3.8RR477 pKa = 11.84GFSEE481 pKa = 4.4GWWWVSALGAKK492 pKa = 9.14TMVGLHH498 pKa = 7.18PIPGYY503 pKa = 9.96PPLCQRR509 pKa = 11.84DD510 pKa = 4.09KK511 pKa = 11.37LWVFRR516 pKa = 11.84NPSAFPLWAVVSHH529 pKa = 6.77LPTPGEE535 pKa = 4.0LPVPAGEE542 pKa = 4.74AVLKK546 pKa = 10.45GQGRR550 pKa = 11.84TSWQFQVRR558 pKa = 11.84AQTNGVFLWLFAPDD572 pKa = 3.96PGWRR576 pKa = 11.84FVVDD580 pKa = 4.12GKK582 pKa = 10.46KK583 pKa = 10.26VKK585 pKa = 9.85PIRR588 pKa = 11.84GAGILQGVPVAAGDD602 pKa = 3.64HH603 pKa = 5.49EE604 pKa = 4.61VRR606 pKa = 11.84VVYY609 pKa = 10.3RR610 pKa = 11.84PPGLMAGLVVSLASLFLLGVLWRR633 pKa = 11.84RR634 pKa = 3.6
MM1 pKa = 7.5SFRR4 pKa = 11.84HH5 pKa = 5.57AFSLAVLPGLALVLASFAGWVPAARR30 pKa = 11.84DD31 pKa = 3.48NPTYY35 pKa = 10.19FVPLRR40 pKa = 11.84AQLARR45 pKa = 11.84VFTGEE50 pKa = 3.76ASPWLNPQVGCGEE63 pKa = 4.18PFFANPQSALLYY75 pKa = 8.84PPAWGALLLPPEE87 pKa = 3.99QAIGVEE93 pKa = 4.37VGLHH97 pKa = 6.2LMLLALGVARR107 pKa = 11.84LARR110 pKa = 11.84RR111 pKa = 11.84LGGTPTGSLAAAWGAALSGPVLSSAGMLNNLEE143 pKa = 4.06TAAWLPWVWDD153 pKa = 3.56AALSGRR159 pKa = 11.84FGVLALTVTGSFLAAEE175 pKa = 4.43PVLALLGAAGAVWLLPRR192 pKa = 11.84WQAVRR197 pKa = 11.84AVLLGFALCSVQALPMAFWIAGGDD221 pKa = 3.6RR222 pKa = 11.84GPDD225 pKa = 3.21KK226 pKa = 10.74PLEE229 pKa = 4.25AVSLGGVSLGEE240 pKa = 4.06LPALVVPGFPLPPVEE255 pKa = 5.03VRR257 pKa = 11.84FLPIIALPLWLFLALGSLRR276 pKa = 11.84RR277 pKa = 11.84GEE279 pKa = 4.14TARR282 pKa = 11.84TRR284 pKa = 11.84LASLAAVFTFLAVLPALPWGDD305 pKa = 4.04ALWAGLSFGLVRR317 pKa = 11.84LPGRR321 pKa = 11.84FLIPATVALAAVAGSGKK338 pKa = 10.17LPQSRR343 pKa = 11.84KK344 pKa = 7.14WVATALGLGAVGAVVSRR361 pKa = 11.84EE362 pKa = 3.59PWLAAGQGVLAALAPWGMGWAAAGSLFLATYY393 pKa = 7.4TVPVLQLQKK402 pKa = 10.15WKK404 pKa = 9.96PEE406 pKa = 3.8PVLCLSAQSAGRR418 pKa = 11.84LYY420 pKa = 9.77PLPVDD425 pKa = 4.43GVQLRR430 pKa = 11.84WTLEE434 pKa = 3.61RR435 pKa = 11.84GTRR438 pKa = 11.84GAASLGWGYY447 pKa = 11.33SVLLDD452 pKa = 3.53GRR454 pKa = 11.84SLARR458 pKa = 11.84SFAPVTNRR466 pKa = 11.84ALAQHH471 pKa = 7.25LAQADD476 pKa = 3.8RR477 pKa = 11.84GFSEE481 pKa = 4.4GWWWVSALGAKK492 pKa = 9.14TMVGLHH498 pKa = 7.18PIPGYY503 pKa = 9.96PPLCQRR509 pKa = 11.84DD510 pKa = 4.09KK511 pKa = 11.37LWVFRR516 pKa = 11.84NPSAFPLWAVVSHH529 pKa = 6.77LPTPGEE535 pKa = 4.0LPVPAGEE542 pKa = 4.74AVLKK546 pKa = 10.45GQGRR550 pKa = 11.84TSWQFQVRR558 pKa = 11.84AQTNGVFLWLFAPDD572 pKa = 3.96PGWRR576 pKa = 11.84FVVDD580 pKa = 4.12GKK582 pKa = 10.46KK583 pKa = 10.26VKK585 pKa = 9.85PIRR588 pKa = 11.84GAGILQGVPVAAGDD602 pKa = 3.64HH603 pKa = 5.49EE604 pKa = 4.61VRR606 pKa = 11.84VVYY609 pKa = 10.3RR610 pKa = 11.84PPGLMAGLVVSLASLFLLGVLWRR633 pKa = 11.84RR634 pKa = 3.6
Molecular weight: 67.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
781730 |
49 |
1332 |
347.1 |
37.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.564 ± 0.063 | 0.939 ± 0.02 |
3.995 ± 0.035 | 6.877 ± 0.058 |
4.051 ± 0.037 | 8.378 ± 0.044 |
1.979 ± 0.024 | 3.183 ± 0.035 |
3.404 ± 0.035 | 12.106 ± 0.075 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.631 ± 0.019 | 2.102 ± 0.029 |
6.09 ± 0.042 | 3.807 ± 0.029 |
7.379 ± 0.047 | 5.046 ± 0.04 |
4.641 ± 0.04 | 8.976 ± 0.045 |
1.716 ± 0.029 | 2.135 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
