
Butyrivibrio hungatei
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Butyrivibrio
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
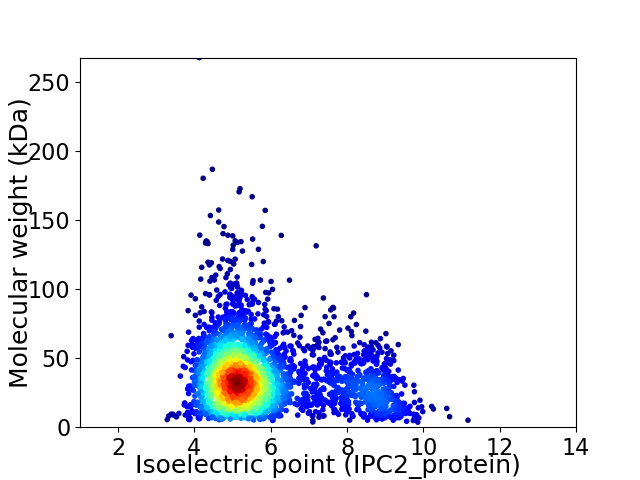
Virtual 2D-PAGE plot for 3003 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
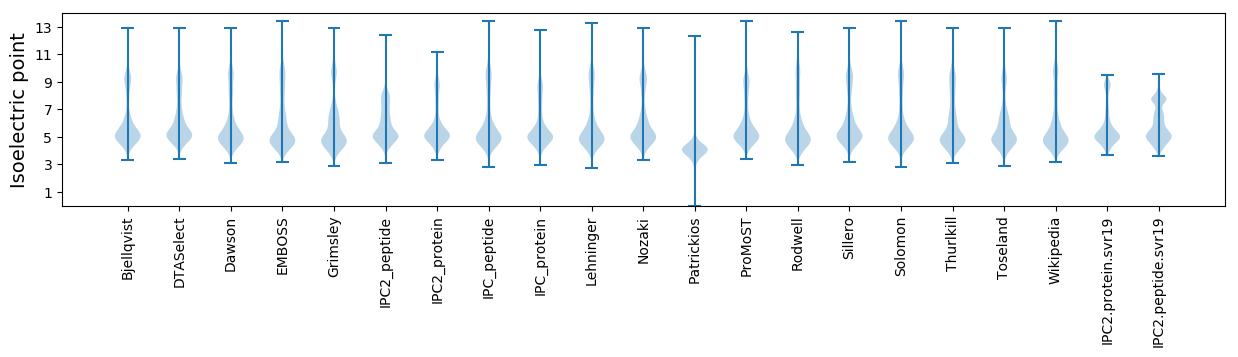
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D9NYN1|A0A1D9NYN1_9FIRM RNA polymerase sigma factor sigma-70 family OS=Butyrivibrio hungatei OX=185008 GN=bhn_I0328 PE=3 SV=1
MM1 pKa = 7.15STHH4 pKa = 6.61KK5 pKa = 10.86LIDD8 pKa = 3.55KK9 pKa = 10.05VCIFVIIFAVLATVLFMNGSAIGITADD36 pKa = 3.76DD37 pKa = 4.19SVEE40 pKa = 3.84MGYY43 pKa = 10.74EE44 pKa = 3.75EE45 pKa = 6.05LLFDD49 pKa = 4.83DD50 pKa = 5.48SYY52 pKa = 12.03VHH54 pKa = 6.9TIDD57 pKa = 5.6IEE59 pKa = 4.59IDD61 pKa = 3.3DD62 pKa = 4.18WDD64 pKa = 4.32SFLEE68 pKa = 4.25TATSEE73 pKa = 4.44EE74 pKa = 4.24YY75 pKa = 10.59SAANVTIDD83 pKa = 3.61GEE85 pKa = 4.59TVYY88 pKa = 11.19NVGIRR93 pKa = 11.84GKK95 pKa = 10.95GNTSLSTVEE104 pKa = 4.34QLDD107 pKa = 3.37SDD109 pKa = 4.04RR110 pKa = 11.84YY111 pKa = 8.64SFKK114 pKa = 10.7IEE116 pKa = 3.73FDD118 pKa = 3.97CYY120 pKa = 10.23EE121 pKa = 4.1SSNTYY126 pKa = 10.17HH127 pKa = 7.22GLDD130 pKa = 3.66KK131 pKa = 11.32LCLNNLIQDD140 pKa = 3.77YY141 pKa = 9.67TMMKK145 pKa = 10.41DD146 pKa = 3.3YY147 pKa = 10.95LAYY150 pKa = 10.07KK151 pKa = 10.57LMGEE155 pKa = 4.66FGVDD159 pKa = 3.43APLVSYY165 pKa = 10.13VYY167 pKa = 9.28ITVNGEE173 pKa = 3.55DD174 pKa = 3.09WGLYY178 pKa = 9.37LAVEE182 pKa = 4.19AVEE185 pKa = 5.54DD186 pKa = 4.34SFLEE190 pKa = 4.32RR191 pKa = 11.84NNGTNYY197 pKa = 10.5GNLYY201 pKa = 10.55KK202 pKa = 10.15PDD204 pKa = 3.97TLSMGGGRR212 pKa = 11.84GNGRR216 pKa = 11.84DD217 pKa = 3.46FDD219 pKa = 4.39MNDD222 pKa = 3.06VDD224 pKa = 5.2SSDD227 pKa = 3.55MPEE230 pKa = 4.36GFGKK234 pKa = 10.71KK235 pKa = 9.79PGDD238 pKa = 3.45RR239 pKa = 11.84SGEE242 pKa = 4.03LPGKK246 pKa = 9.82DD247 pKa = 3.49QEE249 pKa = 4.48EE250 pKa = 4.31HH251 pKa = 6.94GGMGSDD257 pKa = 4.23DD258 pKa = 4.32SKK260 pKa = 11.54LKK262 pKa = 11.14YY263 pKa = 9.89IDD265 pKa = 5.48DD266 pKa = 5.91DD267 pKa = 3.62IDD269 pKa = 3.95SYY271 pKa = 12.23SNIFDD276 pKa = 3.75SAKK279 pKa = 10.3TNIKK283 pKa = 10.49KK284 pKa = 9.98SDD286 pKa = 3.42KK287 pKa = 10.37KK288 pKa = 10.98RR289 pKa = 11.84LINSLKK295 pKa = 9.22TLSEE299 pKa = 4.61GIEE302 pKa = 3.95EE303 pKa = 3.92QDD305 pKa = 2.98ADD307 pKa = 4.48KK308 pKa = 10.83IAEE311 pKa = 4.3AVDD314 pKa = 3.13VDD316 pKa = 3.37AVMRR320 pKa = 11.84YY321 pKa = 7.49MVVHH325 pKa = 6.34NFLVNGDD332 pKa = 4.21SYY334 pKa = 10.44TGSMIHH340 pKa = 6.56NYY342 pKa = 9.82YY343 pKa = 10.51LYY345 pKa = 10.82EE346 pKa = 4.33EE347 pKa = 5.53DD348 pKa = 5.12GLLSMIPWDD357 pKa = 3.61YY358 pKa = 11.46NLAYY362 pKa = 9.32GTFQGQDD369 pKa = 2.49ADD371 pKa = 4.13GAVNDD376 pKa = 5.66PIDD379 pKa = 3.87TPLSVSEE386 pKa = 5.06DD387 pKa = 3.39SDD389 pKa = 3.91DD390 pKa = 4.37RR391 pKa = 11.84PMFSWITSCDD401 pKa = 3.57EE402 pKa = 3.91YY403 pKa = 11.15TEE405 pKa = 4.72MYY407 pKa = 9.56HH408 pKa = 7.2EE409 pKa = 4.81YY410 pKa = 10.71YY411 pKa = 11.03ADD413 pKa = 3.7FLEE416 pKa = 4.05QFYY419 pKa = 10.46TSGYY423 pKa = 9.15LQDD426 pKa = 5.61LIEE429 pKa = 4.38STYY432 pKa = 11.85DD433 pKa = 3.28MISEE437 pKa = 4.24YY438 pKa = 10.61VEE440 pKa = 4.02KK441 pKa = 10.73DD442 pKa = 2.98PTKK445 pKa = 10.56FCTTEE450 pKa = 3.74EE451 pKa = 4.29FEE453 pKa = 4.61TAVEE457 pKa = 4.19TMKK460 pKa = 10.75TFVEE464 pKa = 4.69LRR466 pKa = 11.84CQSISGQLDD475 pKa = 3.42GTIGSTDD482 pKa = 3.28AEE484 pKa = 4.25QEE486 pKa = 4.27EE487 pKa = 4.69NPDD490 pKa = 3.68ALIDD494 pKa = 3.97ASSITLSDD502 pKa = 3.68MGSMGGGMGGGPGQGDD518 pKa = 4.45GEE520 pKa = 4.44HH521 pKa = 6.66SGPPSGDD528 pKa = 3.2PDD530 pKa = 3.73QNSDD534 pKa = 3.13GG535 pKa = 3.99
MM1 pKa = 7.15STHH4 pKa = 6.61KK5 pKa = 10.86LIDD8 pKa = 3.55KK9 pKa = 10.05VCIFVIIFAVLATVLFMNGSAIGITADD36 pKa = 3.76DD37 pKa = 4.19SVEE40 pKa = 3.84MGYY43 pKa = 10.74EE44 pKa = 3.75EE45 pKa = 6.05LLFDD49 pKa = 4.83DD50 pKa = 5.48SYY52 pKa = 12.03VHH54 pKa = 6.9TIDD57 pKa = 5.6IEE59 pKa = 4.59IDD61 pKa = 3.3DD62 pKa = 4.18WDD64 pKa = 4.32SFLEE68 pKa = 4.25TATSEE73 pKa = 4.44EE74 pKa = 4.24YY75 pKa = 10.59SAANVTIDD83 pKa = 3.61GEE85 pKa = 4.59TVYY88 pKa = 11.19NVGIRR93 pKa = 11.84GKK95 pKa = 10.95GNTSLSTVEE104 pKa = 4.34QLDD107 pKa = 3.37SDD109 pKa = 4.04RR110 pKa = 11.84YY111 pKa = 8.64SFKK114 pKa = 10.7IEE116 pKa = 3.73FDD118 pKa = 3.97CYY120 pKa = 10.23EE121 pKa = 4.1SSNTYY126 pKa = 10.17HH127 pKa = 7.22GLDD130 pKa = 3.66KK131 pKa = 11.32LCLNNLIQDD140 pKa = 3.77YY141 pKa = 9.67TMMKK145 pKa = 10.41DD146 pKa = 3.3YY147 pKa = 10.95LAYY150 pKa = 10.07KK151 pKa = 10.57LMGEE155 pKa = 4.66FGVDD159 pKa = 3.43APLVSYY165 pKa = 10.13VYY167 pKa = 9.28ITVNGEE173 pKa = 3.55DD174 pKa = 3.09WGLYY178 pKa = 9.37LAVEE182 pKa = 4.19AVEE185 pKa = 5.54DD186 pKa = 4.34SFLEE190 pKa = 4.32RR191 pKa = 11.84NNGTNYY197 pKa = 10.5GNLYY201 pKa = 10.55KK202 pKa = 10.15PDD204 pKa = 3.97TLSMGGGRR212 pKa = 11.84GNGRR216 pKa = 11.84DD217 pKa = 3.46FDD219 pKa = 4.39MNDD222 pKa = 3.06VDD224 pKa = 5.2SSDD227 pKa = 3.55MPEE230 pKa = 4.36GFGKK234 pKa = 10.71KK235 pKa = 9.79PGDD238 pKa = 3.45RR239 pKa = 11.84SGEE242 pKa = 4.03LPGKK246 pKa = 9.82DD247 pKa = 3.49QEE249 pKa = 4.48EE250 pKa = 4.31HH251 pKa = 6.94GGMGSDD257 pKa = 4.23DD258 pKa = 4.32SKK260 pKa = 11.54LKK262 pKa = 11.14YY263 pKa = 9.89IDD265 pKa = 5.48DD266 pKa = 5.91DD267 pKa = 3.62IDD269 pKa = 3.95SYY271 pKa = 12.23SNIFDD276 pKa = 3.75SAKK279 pKa = 10.3TNIKK283 pKa = 10.49KK284 pKa = 9.98SDD286 pKa = 3.42KK287 pKa = 10.37KK288 pKa = 10.98RR289 pKa = 11.84LINSLKK295 pKa = 9.22TLSEE299 pKa = 4.61GIEE302 pKa = 3.95EE303 pKa = 3.92QDD305 pKa = 2.98ADD307 pKa = 4.48KK308 pKa = 10.83IAEE311 pKa = 4.3AVDD314 pKa = 3.13VDD316 pKa = 3.37AVMRR320 pKa = 11.84YY321 pKa = 7.49MVVHH325 pKa = 6.34NFLVNGDD332 pKa = 4.21SYY334 pKa = 10.44TGSMIHH340 pKa = 6.56NYY342 pKa = 9.82YY343 pKa = 10.51LYY345 pKa = 10.82EE346 pKa = 4.33EE347 pKa = 5.53DD348 pKa = 5.12GLLSMIPWDD357 pKa = 3.61YY358 pKa = 11.46NLAYY362 pKa = 9.32GTFQGQDD369 pKa = 2.49ADD371 pKa = 4.13GAVNDD376 pKa = 5.66PIDD379 pKa = 3.87TPLSVSEE386 pKa = 5.06DD387 pKa = 3.39SDD389 pKa = 3.91DD390 pKa = 4.37RR391 pKa = 11.84PMFSWITSCDD401 pKa = 3.57EE402 pKa = 3.91YY403 pKa = 11.15TEE405 pKa = 4.72MYY407 pKa = 9.56HH408 pKa = 7.2EE409 pKa = 4.81YY410 pKa = 10.71YY411 pKa = 11.03ADD413 pKa = 3.7FLEE416 pKa = 4.05QFYY419 pKa = 10.46TSGYY423 pKa = 9.15LQDD426 pKa = 5.61LIEE429 pKa = 4.38STYY432 pKa = 11.85DD433 pKa = 3.28MISEE437 pKa = 4.24YY438 pKa = 10.61VEE440 pKa = 4.02KK441 pKa = 10.73DD442 pKa = 2.98PTKK445 pKa = 10.56FCTTEE450 pKa = 3.74EE451 pKa = 4.29FEE453 pKa = 4.61TAVEE457 pKa = 4.19TMKK460 pKa = 10.75TFVEE464 pKa = 4.69LRR466 pKa = 11.84CQSISGQLDD475 pKa = 3.42GTIGSTDD482 pKa = 3.28AEE484 pKa = 4.25QEE486 pKa = 4.27EE487 pKa = 4.69NPDD490 pKa = 3.68ALIDD494 pKa = 3.97ASSITLSDD502 pKa = 3.68MGSMGGGMGGGPGQGDD518 pKa = 4.45GEE520 pKa = 4.44HH521 pKa = 6.66SGPPSGDD528 pKa = 3.2PDD530 pKa = 3.73QNSDD534 pKa = 3.13GG535 pKa = 3.99
Molecular weight: 59.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D9P5J9|A0A1D9P5J9_9FIRM Sortase family protein OS=Butyrivibrio hungatei OX=185008 GN=bhn_II020 PE=4 SV=1
MM1 pKa = 7.58KK2 pKa = 8.74MTFQPHH8 pKa = 5.93KK9 pKa = 8.68LQRR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.36GGRR28 pKa = 11.84KK29 pKa = 8.97VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.58KK2 pKa = 8.74MTFQPHH8 pKa = 5.93KK9 pKa = 8.68LQRR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.36GGRR28 pKa = 11.84KK29 pKa = 8.97VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1018659 |
29 |
2445 |
339.2 |
38.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.199 ± 0.04 | 1.368 ± 0.019 |
6.509 ± 0.044 | 7.304 ± 0.05 |
4.412 ± 0.036 | 6.827 ± 0.038 |
1.612 ± 0.018 | 7.78 ± 0.043 |
7.277 ± 0.038 | 8.46 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.027 ± 0.023 | 4.771 ± 0.028 |
3.091 ± 0.026 | 2.648 ± 0.024 |
3.797 ± 0.027 | 6.233 ± 0.042 |
5.34 ± 0.039 | 6.956 ± 0.032 |
0.883 ± 0.015 | 4.506 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
