
Micavibrio aeruginosavorus (strain ARL-13)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Alphaproteobacteria incertae sedis; Micavibrio; Micavibrio aeruginosavorus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
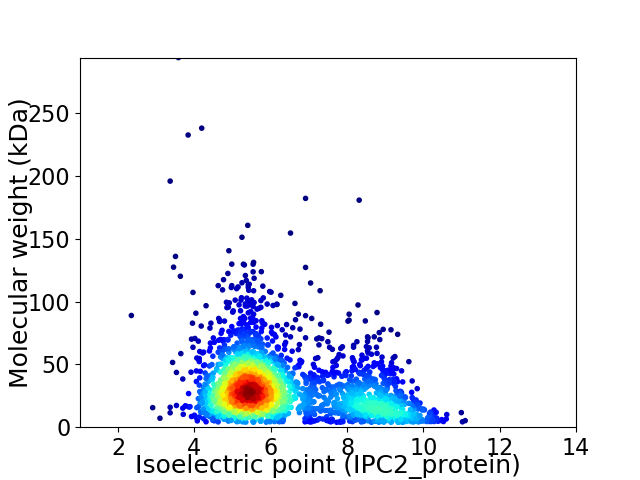
Virtual 2D-PAGE plot for 2432 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G2KMQ4|G2KMQ4_MICAA Methyltransferase small domain protein OS=Micavibrio aeruginosavorus (strain ARL-13) OX=856793 GN=MICA_94 PE=4 SV=1
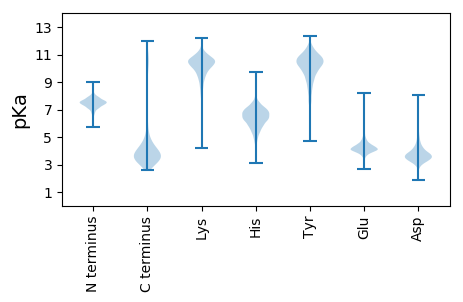
MM1 pKa = 7.54CAGAKK6 pKa = 9.75ASGMADD12 pKa = 3.21TVFTGTASGDD22 pKa = 3.62FMVMQGDD29 pKa = 4.28LQHH32 pKa = 5.78MTMTLTNPYY41 pKa = 9.19TGEE44 pKa = 4.08TVNINDD50 pKa = 3.55VYY52 pKa = 11.02KK53 pKa = 11.02VNNAAYY59 pKa = 9.43DD60 pKa = 3.9GLGGTDD66 pKa = 3.48ALLMSEE72 pKa = 4.8HH73 pKa = 7.33GDD75 pKa = 3.21MLLIKK80 pKa = 10.51DD81 pKa = 3.81ADD83 pKa = 3.71GDD85 pKa = 3.89QTLKK89 pKa = 11.01NIEE92 pKa = 4.05QIIAGDD98 pKa = 3.72GGDD101 pKa = 3.97VIVLADD107 pKa = 4.6AEE109 pKa = 4.27IQYY112 pKa = 11.32GNVAIDD118 pKa = 3.54GGGSGDD124 pKa = 4.95IIWSNNGNDD133 pKa = 4.08TIFALDD139 pKa = 3.74GNDD142 pKa = 3.17IVDD145 pKa = 4.02GGGGLDD151 pKa = 4.12KK152 pKa = 10.61IYY154 pKa = 10.82GGWGSDD160 pKa = 3.46YY161 pKa = 11.13LRR163 pKa = 11.84GGNGDD168 pKa = 4.17DD169 pKa = 3.93IIYY172 pKa = 10.49GGSRR176 pKa = 11.84SDD178 pKa = 3.59TEE180 pKa = 4.6AEE182 pKa = 4.05YY183 pKa = 8.5HH184 pKa = 5.64TVSTYY189 pKa = 10.92EE190 pKa = 3.72KK191 pKa = 9.61FFTVTHH197 pKa = 6.45SFPTLNEE204 pKa = 4.15GVQITPQQQAYY215 pKa = 9.74LGYY218 pKa = 9.3PSGEE222 pKa = 4.27TIVTFTTTATLKK234 pKa = 10.56FVSTDD239 pKa = 2.81AGYY242 pKa = 10.83KK243 pKa = 8.4NTVGVYY249 pKa = 8.97TVDD252 pKa = 3.11PDD254 pKa = 3.7GTIRR258 pKa = 11.84NADD261 pKa = 3.71FAFMNARR268 pKa = 11.84ATASGTSHH276 pKa = 6.12EE277 pKa = 4.8FEE279 pKa = 4.15VSGAVGVQIGLFMIANGYY297 pKa = 8.12NANGGYY303 pKa = 10.51GGIDD307 pKa = 3.59MNTGTLNFYY316 pKa = 10.88YY317 pKa = 10.45NYY319 pKa = 10.3GQADD323 pKa = 3.4QRR325 pKa = 11.84LAKK328 pKa = 9.73VTDD331 pKa = 4.25DD332 pKa = 4.04GDD334 pKa = 3.95NVSLIWSDD342 pKa = 3.09GTTDD346 pKa = 3.48KK347 pKa = 11.34LIAGTIYY354 pKa = 10.47HH355 pKa = 6.65SSASDD360 pKa = 3.46GSSSLNRR367 pKa = 11.84DD368 pKa = 3.58DD369 pKa = 5.06NIHH372 pKa = 6.52VITGLANAGDD382 pKa = 4.13NTTLRR387 pKa = 11.84LGFEE391 pKa = 4.64DD392 pKa = 4.49QDD394 pKa = 3.89HH395 pKa = 7.05LGDD398 pKa = 4.28ADD400 pKa = 4.13FNDD403 pKa = 4.37VIVDD407 pKa = 4.05LQVEE411 pKa = 4.5TQVVEE416 pKa = 4.77TILRR420 pKa = 11.84PDD422 pKa = 3.98DD423 pKa = 5.12DD424 pKa = 4.18ILYY427 pKa = 10.71GDD429 pKa = 4.85AGNDD433 pKa = 3.3TLYY436 pKa = 11.24GGFGNDD442 pKa = 3.37VLNGGTGSDD451 pKa = 3.7HH452 pKa = 7.41LYY454 pKa = 10.72GGQGADD460 pKa = 2.81TFVFDD465 pKa = 4.9NMDD468 pKa = 3.49GFVDD472 pKa = 4.17TVHH475 pKa = 7.16DD476 pKa = 5.25FKK478 pKa = 11.49ASEE481 pKa = 4.25GDD483 pKa = 3.54VLDD486 pKa = 4.41ISDD489 pKa = 4.14ILSGYY494 pKa = 9.71DD495 pKa = 3.64AGVDD499 pKa = 4.71DD500 pKa = 4.56ITDD503 pKa = 3.77FLKK506 pKa = 10.8FVQNGSNVDD515 pKa = 3.75LYY517 pKa = 11.96VNADD521 pKa = 3.54GDD523 pKa = 3.93SGGAFDD529 pKa = 4.47RR530 pKa = 11.84VAVIVGGVGGASVQDD545 pKa = 4.33LLDD548 pKa = 4.02GGAIVAAQTAVV559 pKa = 2.85
MM1 pKa = 7.54CAGAKK6 pKa = 9.75ASGMADD12 pKa = 3.21TVFTGTASGDD22 pKa = 3.62FMVMQGDD29 pKa = 4.28LQHH32 pKa = 5.78MTMTLTNPYY41 pKa = 9.19TGEE44 pKa = 4.08TVNINDD50 pKa = 3.55VYY52 pKa = 11.02KK53 pKa = 11.02VNNAAYY59 pKa = 9.43DD60 pKa = 3.9GLGGTDD66 pKa = 3.48ALLMSEE72 pKa = 4.8HH73 pKa = 7.33GDD75 pKa = 3.21MLLIKK80 pKa = 10.51DD81 pKa = 3.81ADD83 pKa = 3.71GDD85 pKa = 3.89QTLKK89 pKa = 11.01NIEE92 pKa = 4.05QIIAGDD98 pKa = 3.72GGDD101 pKa = 3.97VIVLADD107 pKa = 4.6AEE109 pKa = 4.27IQYY112 pKa = 11.32GNVAIDD118 pKa = 3.54GGGSGDD124 pKa = 4.95IIWSNNGNDD133 pKa = 4.08TIFALDD139 pKa = 3.74GNDD142 pKa = 3.17IVDD145 pKa = 4.02GGGGLDD151 pKa = 4.12KK152 pKa = 10.61IYY154 pKa = 10.82GGWGSDD160 pKa = 3.46YY161 pKa = 11.13LRR163 pKa = 11.84GGNGDD168 pKa = 4.17DD169 pKa = 3.93IIYY172 pKa = 10.49GGSRR176 pKa = 11.84SDD178 pKa = 3.59TEE180 pKa = 4.6AEE182 pKa = 4.05YY183 pKa = 8.5HH184 pKa = 5.64TVSTYY189 pKa = 10.92EE190 pKa = 3.72KK191 pKa = 9.61FFTVTHH197 pKa = 6.45SFPTLNEE204 pKa = 4.15GVQITPQQQAYY215 pKa = 9.74LGYY218 pKa = 9.3PSGEE222 pKa = 4.27TIVTFTTTATLKK234 pKa = 10.56FVSTDD239 pKa = 2.81AGYY242 pKa = 10.83KK243 pKa = 8.4NTVGVYY249 pKa = 8.97TVDD252 pKa = 3.11PDD254 pKa = 3.7GTIRR258 pKa = 11.84NADD261 pKa = 3.71FAFMNARR268 pKa = 11.84ATASGTSHH276 pKa = 6.12EE277 pKa = 4.8FEE279 pKa = 4.15VSGAVGVQIGLFMIANGYY297 pKa = 8.12NANGGYY303 pKa = 10.51GGIDD307 pKa = 3.59MNTGTLNFYY316 pKa = 10.88YY317 pKa = 10.45NYY319 pKa = 10.3GQADD323 pKa = 3.4QRR325 pKa = 11.84LAKK328 pKa = 9.73VTDD331 pKa = 4.25DD332 pKa = 4.04GDD334 pKa = 3.95NVSLIWSDD342 pKa = 3.09GTTDD346 pKa = 3.48KK347 pKa = 11.34LIAGTIYY354 pKa = 10.47HH355 pKa = 6.65SSASDD360 pKa = 3.46GSSSLNRR367 pKa = 11.84DD368 pKa = 3.58DD369 pKa = 5.06NIHH372 pKa = 6.52VITGLANAGDD382 pKa = 4.13NTTLRR387 pKa = 11.84LGFEE391 pKa = 4.64DD392 pKa = 4.49QDD394 pKa = 3.89HH395 pKa = 7.05LGDD398 pKa = 4.28ADD400 pKa = 4.13FNDD403 pKa = 4.37VIVDD407 pKa = 4.05LQVEE411 pKa = 4.5TQVVEE416 pKa = 4.77TILRR420 pKa = 11.84PDD422 pKa = 3.98DD423 pKa = 5.12DD424 pKa = 4.18ILYY427 pKa = 10.71GDD429 pKa = 4.85AGNDD433 pKa = 3.3TLYY436 pKa = 11.24GGFGNDD442 pKa = 3.37VLNGGTGSDD451 pKa = 3.7HH452 pKa = 7.41LYY454 pKa = 10.72GGQGADD460 pKa = 2.81TFVFDD465 pKa = 4.9NMDD468 pKa = 3.49GFVDD472 pKa = 4.17TVHH475 pKa = 7.16DD476 pKa = 5.25FKK478 pKa = 11.49ASEE481 pKa = 4.25GDD483 pKa = 3.54VLDD486 pKa = 4.41ISDD489 pKa = 4.14ILSGYY494 pKa = 9.71DD495 pKa = 3.64AGVDD499 pKa = 4.71DD500 pKa = 4.56ITDD503 pKa = 3.77FLKK506 pKa = 10.8FVQNGSNVDD515 pKa = 3.75LYY517 pKa = 11.96VNADD521 pKa = 3.54GDD523 pKa = 3.93SGGAFDD529 pKa = 4.47RR530 pKa = 11.84VAVIVGGVGGASVQDD545 pKa = 4.33LLDD548 pKa = 4.02GGAIVAAQTAVV559 pKa = 2.85
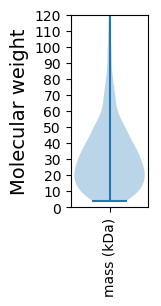
Molecular weight: 58.57 kDa
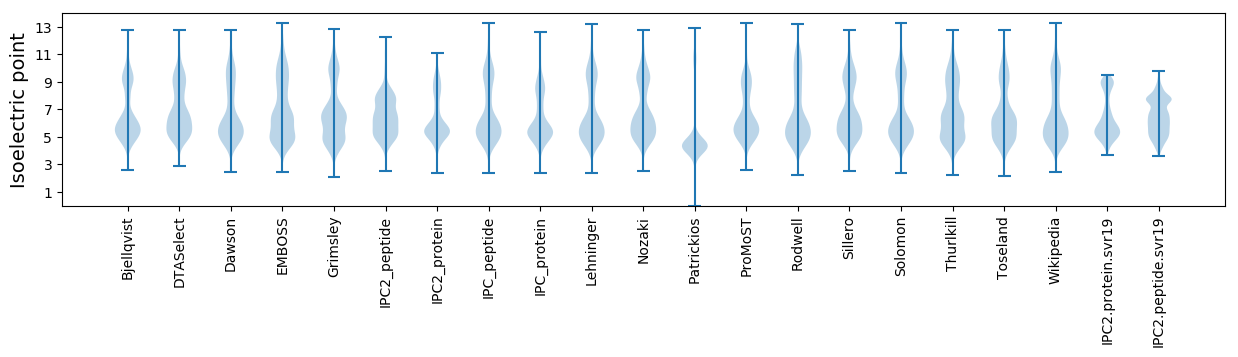
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G2KS99|G2KS99_MICAA HemY family protein OS=Micavibrio aeruginosavorus (strain ARL-13) OX=856793 GN=MICA_439 PE=4 SV=1
MM1 pKa = 7.17WRR3 pKa = 11.84AINARR8 pKa = 11.84HH9 pKa = 5.99IKK11 pKa = 9.85ILPIPKK17 pKa = 9.27RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.83SGPILLRR28 pKa = 11.84GSARR32 pKa = 11.84APPAVVV38 pKa = 2.92
MM1 pKa = 7.17WRR3 pKa = 11.84AINARR8 pKa = 11.84HH9 pKa = 5.99IKK11 pKa = 9.85ILPIPKK17 pKa = 9.27RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.83SGPILLRR28 pKa = 11.84GSARR32 pKa = 11.84APPAVVV38 pKa = 2.92
Molecular weight: 4.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
743786 |
37 |
2892 |
305.8 |
33.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.746 ± 0.059 | 0.917 ± 0.019 |
6.612 ± 0.051 | 5.285 ± 0.05 |
3.728 ± 0.036 | 8.038 ± 0.071 |
2.202 ± 0.025 | 5.862 ± 0.036 |
4.613 ± 0.055 | 8.955 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.214 ± 0.029 | 3.762 ± 0.049 |
4.824 ± 0.039 | 3.506 ± 0.028 |
5.825 ± 0.058 | 5.37 ± 0.047 |
5.84 ± 0.052 | 6.986 ± 0.043 |
1.116 ± 0.02 | 2.6 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
