
Gemmiger formicilis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriales incertae sedis; Gemmiger
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
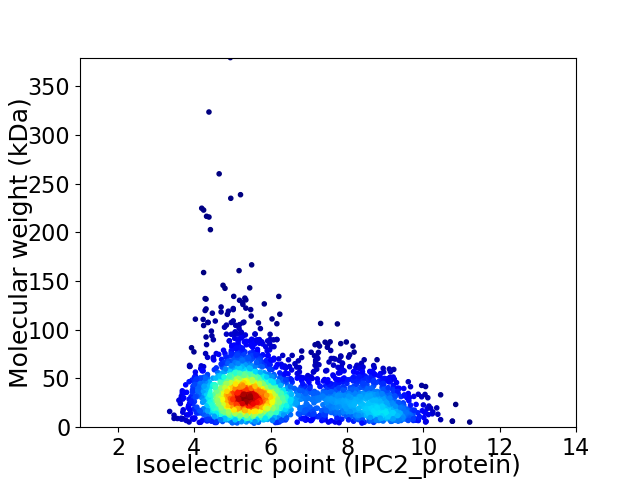
Virtual 2D-PAGE plot for 2819 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1T4X083|A0A1T4X083_9FIRM Predicted transcriptional regulator OS=Gemmiger formicilis OX=745368 GN=SAMN02745178_01222 PE=3 SV=1
MM1 pKa = 7.49KK2 pKa = 10.22LKK4 pKa = 10.57NFRR7 pKa = 11.84RR8 pKa = 11.84VAALGMAVGMTAVLAACGNTADD30 pKa = 4.25TASSEE35 pKa = 4.28AAEE38 pKa = 4.52STAEE42 pKa = 4.04TADD45 pKa = 3.63TAEE48 pKa = 4.22EE49 pKa = 4.16ASAEE53 pKa = 4.25TADD56 pKa = 4.76AEE58 pKa = 4.43YY59 pKa = 10.3AYY61 pKa = 10.92LADD64 pKa = 5.2FSFSDD69 pKa = 3.63AFDD72 pKa = 3.6EE73 pKa = 4.2NGYY76 pKa = 10.6LKK78 pKa = 10.8GVTAADD84 pKa = 4.18YY85 pKa = 8.63VTLPDD90 pKa = 4.93DD91 pKa = 3.95YY92 pKa = 12.13ADD94 pKa = 3.47ITISADD100 pKa = 3.12LGQVSDD106 pKa = 5.55DD107 pKa = 5.35DD108 pKa = 4.2INDD111 pKa = 4.21YY112 pKa = 10.91IDD114 pKa = 4.99SNVLSNFATTNEE126 pKa = 4.19VKK128 pKa = 10.66DD129 pKa = 3.7RR130 pKa = 11.84AAADD134 pKa = 3.66GDD136 pKa = 4.41TVNIDD141 pKa = 3.19YY142 pKa = 10.68VGRR145 pKa = 11.84IDD147 pKa = 5.35GVAFDD152 pKa = 4.87GGDD155 pKa = 3.6TKK157 pKa = 11.39GSGADD162 pKa = 3.45LTLGSGTYY170 pKa = 9.51IDD172 pKa = 4.39NFEE175 pKa = 4.13DD176 pKa = 3.71QIVGHH181 pKa = 5.89TPGEE185 pKa = 4.4TFDD188 pKa = 3.66VTVTFPEE195 pKa = 4.91DD196 pKa = 3.56YY197 pKa = 11.23GNEE200 pKa = 3.96DD201 pKa = 3.92LNGKK205 pKa = 8.37EE206 pKa = 4.38AVFEE210 pKa = 4.27TTLNYY215 pKa = 9.8IKK217 pKa = 10.64EE218 pKa = 4.49SVTPEE223 pKa = 4.03LTDD226 pKa = 3.37DD227 pKa = 3.51WVKK230 pKa = 11.23EE231 pKa = 3.95NLGKK235 pKa = 10.87SMSLNSVDD243 pKa = 3.52EE244 pKa = 4.3LKK246 pKa = 11.21AFVKK250 pKa = 9.79STMLYY255 pKa = 10.59DD256 pKa = 3.66NQASDD261 pKa = 4.32VYY263 pKa = 10.51TALHH267 pKa = 6.85DD268 pKa = 3.85KK269 pKa = 10.96ASYY272 pKa = 10.98ADD274 pKa = 4.5EE275 pKa = 4.72LPQTALDD282 pKa = 3.86YY283 pKa = 11.35YY284 pKa = 11.07RR285 pKa = 11.84DD286 pKa = 3.53VLLYY290 pKa = 10.41RR291 pKa = 11.84IYY293 pKa = 11.12TNAQKK298 pKa = 10.98YY299 pKa = 6.83GTDD302 pKa = 3.38MNALLSSGMMGSSYY316 pKa = 11.3DD317 pKa = 3.75SVDD320 pKa = 4.17AYY322 pKa = 11.24LADD325 pKa = 4.35ANDD328 pKa = 4.27SIKK331 pKa = 10.98SITQQALLMQAVAEE345 pKa = 4.23KK346 pKa = 10.13TGFKK350 pKa = 10.49CDD352 pKa = 3.6TATMNADD359 pKa = 4.03FGRR362 pKa = 11.84YY363 pKa = 9.16YY364 pKa = 10.6GTTDD368 pKa = 3.32PSQYY372 pKa = 10.13VSAYY376 pKa = 8.17GEE378 pKa = 4.36NYY380 pKa = 9.82IKK382 pKa = 10.64MNVLQSDD389 pKa = 3.79VMQNLIDD396 pKa = 3.82NVKK399 pKa = 10.59YY400 pKa = 9.98EE401 pKa = 3.96
MM1 pKa = 7.49KK2 pKa = 10.22LKK4 pKa = 10.57NFRR7 pKa = 11.84RR8 pKa = 11.84VAALGMAVGMTAVLAACGNTADD30 pKa = 4.25TASSEE35 pKa = 4.28AAEE38 pKa = 4.52STAEE42 pKa = 4.04TADD45 pKa = 3.63TAEE48 pKa = 4.22EE49 pKa = 4.16ASAEE53 pKa = 4.25TADD56 pKa = 4.76AEE58 pKa = 4.43YY59 pKa = 10.3AYY61 pKa = 10.92LADD64 pKa = 5.2FSFSDD69 pKa = 3.63AFDD72 pKa = 3.6EE73 pKa = 4.2NGYY76 pKa = 10.6LKK78 pKa = 10.8GVTAADD84 pKa = 4.18YY85 pKa = 8.63VTLPDD90 pKa = 4.93DD91 pKa = 3.95YY92 pKa = 12.13ADD94 pKa = 3.47ITISADD100 pKa = 3.12LGQVSDD106 pKa = 5.55DD107 pKa = 5.35DD108 pKa = 4.2INDD111 pKa = 4.21YY112 pKa = 10.91IDD114 pKa = 4.99SNVLSNFATTNEE126 pKa = 4.19VKK128 pKa = 10.66DD129 pKa = 3.7RR130 pKa = 11.84AAADD134 pKa = 3.66GDD136 pKa = 4.41TVNIDD141 pKa = 3.19YY142 pKa = 10.68VGRR145 pKa = 11.84IDD147 pKa = 5.35GVAFDD152 pKa = 4.87GGDD155 pKa = 3.6TKK157 pKa = 11.39GSGADD162 pKa = 3.45LTLGSGTYY170 pKa = 9.51IDD172 pKa = 4.39NFEE175 pKa = 4.13DD176 pKa = 3.71QIVGHH181 pKa = 5.89TPGEE185 pKa = 4.4TFDD188 pKa = 3.66VTVTFPEE195 pKa = 4.91DD196 pKa = 3.56YY197 pKa = 11.23GNEE200 pKa = 3.96DD201 pKa = 3.92LNGKK205 pKa = 8.37EE206 pKa = 4.38AVFEE210 pKa = 4.27TTLNYY215 pKa = 9.8IKK217 pKa = 10.64EE218 pKa = 4.49SVTPEE223 pKa = 4.03LTDD226 pKa = 3.37DD227 pKa = 3.51WVKK230 pKa = 11.23EE231 pKa = 3.95NLGKK235 pKa = 10.87SMSLNSVDD243 pKa = 3.52EE244 pKa = 4.3LKK246 pKa = 11.21AFVKK250 pKa = 9.79STMLYY255 pKa = 10.59DD256 pKa = 3.66NQASDD261 pKa = 4.32VYY263 pKa = 10.51TALHH267 pKa = 6.85DD268 pKa = 3.85KK269 pKa = 10.96ASYY272 pKa = 10.98ADD274 pKa = 4.5EE275 pKa = 4.72LPQTALDD282 pKa = 3.86YY283 pKa = 11.35YY284 pKa = 11.07RR285 pKa = 11.84DD286 pKa = 3.53VLLYY290 pKa = 10.41RR291 pKa = 11.84IYY293 pKa = 11.12TNAQKK298 pKa = 10.98YY299 pKa = 6.83GTDD302 pKa = 3.38MNALLSSGMMGSSYY316 pKa = 11.3DD317 pKa = 3.75SVDD320 pKa = 4.17AYY322 pKa = 11.24LADD325 pKa = 4.35ANDD328 pKa = 4.27SIKK331 pKa = 10.98SITQQALLMQAVAEE345 pKa = 4.23KK346 pKa = 10.13TGFKK350 pKa = 10.49CDD352 pKa = 3.6TATMNADD359 pKa = 4.03FGRR362 pKa = 11.84YY363 pKa = 9.16YY364 pKa = 10.6GTTDD368 pKa = 3.32PSQYY372 pKa = 10.13VSAYY376 pKa = 8.17GEE378 pKa = 4.36NYY380 pKa = 9.82IKK382 pKa = 10.64MNVLQSDD389 pKa = 3.79VMQNLIDD396 pKa = 3.82NVKK399 pKa = 10.59YY400 pKa = 9.98EE401 pKa = 3.96
Molecular weight: 43.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1T4XSI1|A0A1T4XSI1_9FIRM Uncharacterized protein OS=Gemmiger formicilis OX=745368 GN=SAMN02745178_02227 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.6RR3 pKa = 11.84TFQPKK8 pKa = 8.37KK9 pKa = 7.49RR10 pKa = 11.84QRR12 pKa = 11.84SRR14 pKa = 11.84VHH16 pKa = 6.24GFLNRR21 pKa = 11.84MATKK25 pKa = 10.47NGRR28 pKa = 11.84KK29 pKa = 9.36VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.98GRR39 pKa = 11.84KK40 pKa = 8.89SLTVV44 pKa = 3.12
MM1 pKa = 7.45KK2 pKa = 9.6RR3 pKa = 11.84TFQPKK8 pKa = 8.37KK9 pKa = 7.49RR10 pKa = 11.84QRR12 pKa = 11.84SRR14 pKa = 11.84VHH16 pKa = 6.24GFLNRR21 pKa = 11.84MATKK25 pKa = 10.47NGRR28 pKa = 11.84KK29 pKa = 9.36VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.98GRR39 pKa = 11.84KK40 pKa = 8.89SLTVV44 pKa = 3.12
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
923584 |
39 |
3580 |
327.6 |
36.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.668 ± 0.071 | 1.735 ± 0.021 |
5.86 ± 0.037 | 5.946 ± 0.048 |
3.773 ± 0.035 | 7.517 ± 0.047 |
1.919 ± 0.02 | 5.32 ± 0.043 |
4.844 ± 0.044 | 9.864 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.666 ± 0.026 | 3.593 ± 0.038 |
4.335 ± 0.035 | 3.515 ± 0.029 |
5.305 ± 0.045 | 5.12 ± 0.037 |
6.099 ± 0.052 | 7.011 ± 0.04 |
1.142 ± 0.018 | 3.766 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
