
Folsomia candida (Springtail)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Collembola; Entomobryomorpha; Isotomoidea; Isotomidae; Proisotominae; Folsomia
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
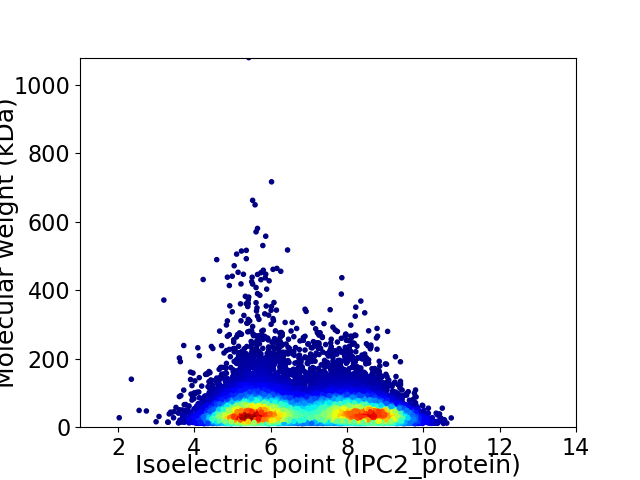
Virtual 2D-PAGE plot for 28565 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A226EJN9|A0A226EJN9_FOLCA Uncharacterized protein OS=Folsomia candida OX=158441 GN=Fcan01_07653 PE=4 SV=1
MM1 pKa = 7.16ACKK4 pKa = 10.01RR5 pKa = 11.84RR6 pKa = 11.84QEE8 pKa = 4.04PDD10 pKa = 2.73TGQKK14 pKa = 10.04RR15 pKa = 11.84SSEE18 pKa = 4.27TGFKK22 pKa = 10.48PSVVWGSGRR31 pKa = 11.84EE32 pKa = 4.1DD33 pKa = 3.11VGGGHH38 pKa = 6.84FLGPVLFFCLFLFFLGPGCGRR59 pKa = 11.84GEE61 pKa = 4.46DD62 pKa = 4.1DD63 pKa = 5.47DD64 pKa = 6.19GGGSGDD70 pKa = 4.44DD71 pKa = 4.55DD72 pKa = 4.84GGSGDD77 pKa = 4.92DD78 pKa = 5.06DD79 pKa = 4.74GGSGDD84 pKa = 4.92DD85 pKa = 5.06DD86 pKa = 4.74GGSGDD91 pKa = 4.92DD92 pKa = 5.06DD93 pKa = 4.74GGSGDD98 pKa = 4.92DD99 pKa = 5.06DD100 pKa = 4.74GGSGDD105 pKa = 4.92DD106 pKa = 5.06DD107 pKa = 4.74GGSGDD112 pKa = 4.92DD113 pKa = 5.06DD114 pKa = 4.74GGSGDD119 pKa = 4.92DD120 pKa = 5.06DD121 pKa = 4.74GGSGDD126 pKa = 4.92DD127 pKa = 5.06DD128 pKa = 4.74GGSGDD133 pKa = 4.92DD134 pKa = 5.06DD135 pKa = 4.74GGSGDD140 pKa = 4.96DD141 pKa = 4.41DD142 pKa = 4.49SGSGGDD148 pKa = 4.49DD149 pKa = 3.47DD150 pKa = 5.93DD151 pKa = 4.88GGRR154 pKa = 11.84GGSSVKK160 pKa = 8.84FTYY163 pKa = 9.73FHH165 pKa = 7.05ISSDD169 pKa = 3.67LKK171 pKa = 11.14SGIVGCGSDD180 pKa = 4.84CCINWLLGLLCCVPTVVHH198 pKa = 6.55ACCFVGEE205 pKa = 4.33EE206 pKa = 4.81DD207 pKa = 4.53IGPPKK212 pKa = 10.5QPVTNVVIVQQPGQPQFHH230 pKa = 6.31VQPQQGGQFQQQCPQAQSPPQYY252 pKa = 11.24VMPQTHH258 pKa = 4.59QQYY261 pKa = 9.83GFKK264 pKa = 10.61YY265 pKa = 10.02
MM1 pKa = 7.16ACKK4 pKa = 10.01RR5 pKa = 11.84RR6 pKa = 11.84QEE8 pKa = 4.04PDD10 pKa = 2.73TGQKK14 pKa = 10.04RR15 pKa = 11.84SSEE18 pKa = 4.27TGFKK22 pKa = 10.48PSVVWGSGRR31 pKa = 11.84EE32 pKa = 4.1DD33 pKa = 3.11VGGGHH38 pKa = 6.84FLGPVLFFCLFLFFLGPGCGRR59 pKa = 11.84GEE61 pKa = 4.46DD62 pKa = 4.1DD63 pKa = 5.47DD64 pKa = 6.19GGGSGDD70 pKa = 4.44DD71 pKa = 4.55DD72 pKa = 4.84GGSGDD77 pKa = 4.92DD78 pKa = 5.06DD79 pKa = 4.74GGSGDD84 pKa = 4.92DD85 pKa = 5.06DD86 pKa = 4.74GGSGDD91 pKa = 4.92DD92 pKa = 5.06DD93 pKa = 4.74GGSGDD98 pKa = 4.92DD99 pKa = 5.06DD100 pKa = 4.74GGSGDD105 pKa = 4.92DD106 pKa = 5.06DD107 pKa = 4.74GGSGDD112 pKa = 4.92DD113 pKa = 5.06DD114 pKa = 4.74GGSGDD119 pKa = 4.92DD120 pKa = 5.06DD121 pKa = 4.74GGSGDD126 pKa = 4.92DD127 pKa = 5.06DD128 pKa = 4.74GGSGDD133 pKa = 4.92DD134 pKa = 5.06DD135 pKa = 4.74GGSGDD140 pKa = 4.96DD141 pKa = 4.41DD142 pKa = 4.49SGSGGDD148 pKa = 4.49DD149 pKa = 3.47DD150 pKa = 5.93DD151 pKa = 4.88GGRR154 pKa = 11.84GGSSVKK160 pKa = 8.84FTYY163 pKa = 9.73FHH165 pKa = 7.05ISSDD169 pKa = 3.67LKK171 pKa = 11.14SGIVGCGSDD180 pKa = 4.84CCINWLLGLLCCVPTVVHH198 pKa = 6.55ACCFVGEE205 pKa = 4.33EE206 pKa = 4.81DD207 pKa = 4.53IGPPKK212 pKa = 10.5QPVTNVVIVQQPGQPQFHH230 pKa = 6.31VQPQQGGQFQQQCPQAQSPPQYY252 pKa = 11.24VMPQTHH258 pKa = 4.59QQYY261 pKa = 9.83GFKK264 pKa = 10.61YY265 pKa = 10.02
Molecular weight: 26.86 kDa
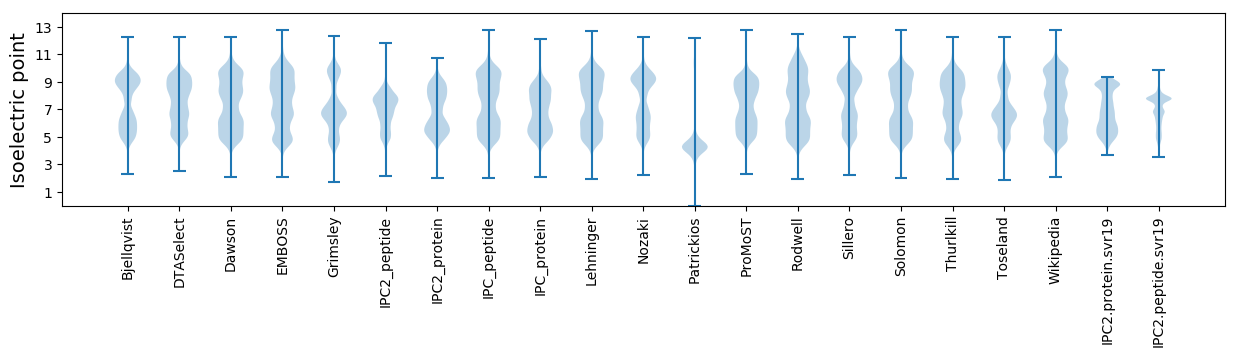
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A226DBG9|A0A226DBG9_FOLCA V(D)J recombination activating protein 1 OS=Folsomia candida OX=158441 GN=Fcan01_22896 PE=4 SV=1
MM1 pKa = 6.92STVRR5 pKa = 11.84PNFSSPPHH13 pKa = 7.01LLIAALLSVQSNSSDD28 pKa = 3.75VIHH31 pKa = 6.79GGSEE35 pKa = 3.9LFSRR39 pKa = 11.84HH40 pKa = 5.0CTTSIGRR47 pKa = 11.84RR48 pKa = 11.84DD49 pKa = 4.15DD50 pKa = 4.06LHH52 pKa = 6.24TLPSILLHH60 pKa = 5.22QFRR63 pKa = 11.84PEE65 pKa = 3.9LVQKK69 pKa = 10.13SIPARR74 pKa = 11.84VHH76 pKa = 5.01TKK78 pKa = 10.2RR79 pKa = 11.84FVMDD83 pKa = 3.73GTVGLAGLIISIPLCNKK100 pKa = 9.17GCSTMFICSGGGGIRR115 pKa = 11.84SNVCKK120 pKa = 10.22FARR123 pKa = 11.84LKK125 pKa = 11.32NNLFSHH131 pKa = 6.68NSHH134 pKa = 5.91VSYY137 pKa = 10.9CIIIVSYY144 pKa = 9.0TFITPNGDD152 pKa = 3.24SVIIIIVVVVQLPLLLRR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84QKK173 pKa = 10.75AAALCKK179 pKa = 9.39SCKK182 pKa = 9.78YY183 pKa = 10.29GQLLRR188 pKa = 11.84TRR190 pKa = 11.84RR191 pKa = 11.84LCNVPMGPASSGRR204 pKa = 11.84QTQASGGRR212 pKa = 11.84NEE214 pKa = 4.5GRR216 pKa = 11.84DD217 pKa = 3.75GSGGGSDD224 pKa = 2.88AGARR228 pKa = 11.84GEE230 pKa = 4.0MSRR233 pKa = 11.84VKK235 pKa = 10.25AGRR238 pKa = 11.84VGWVQWIHH246 pKa = 5.83PSLRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84VSEE255 pKa = 4.39LEE257 pKa = 3.83
MM1 pKa = 6.92STVRR5 pKa = 11.84PNFSSPPHH13 pKa = 7.01LLIAALLSVQSNSSDD28 pKa = 3.75VIHH31 pKa = 6.79GGSEE35 pKa = 3.9LFSRR39 pKa = 11.84HH40 pKa = 5.0CTTSIGRR47 pKa = 11.84RR48 pKa = 11.84DD49 pKa = 4.15DD50 pKa = 4.06LHH52 pKa = 6.24TLPSILLHH60 pKa = 5.22QFRR63 pKa = 11.84PEE65 pKa = 3.9LVQKK69 pKa = 10.13SIPARR74 pKa = 11.84VHH76 pKa = 5.01TKK78 pKa = 10.2RR79 pKa = 11.84FVMDD83 pKa = 3.73GTVGLAGLIISIPLCNKK100 pKa = 9.17GCSTMFICSGGGGIRR115 pKa = 11.84SNVCKK120 pKa = 10.22FARR123 pKa = 11.84LKK125 pKa = 11.32NNLFSHH131 pKa = 6.68NSHH134 pKa = 5.91VSYY137 pKa = 10.9CIIIVSYY144 pKa = 9.0TFITPNGDD152 pKa = 3.24SVIIIIVVVVQLPLLLRR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84QKK173 pKa = 10.75AAALCKK179 pKa = 9.39SCKK182 pKa = 9.78YY183 pKa = 10.29GQLLRR188 pKa = 11.84TRR190 pKa = 11.84RR191 pKa = 11.84LCNVPMGPASSGRR204 pKa = 11.84QTQASGGRR212 pKa = 11.84NEE214 pKa = 4.5GRR216 pKa = 11.84DD217 pKa = 3.75GSGGGSDD224 pKa = 2.88AGARR228 pKa = 11.84GEE230 pKa = 4.0MSRR233 pKa = 11.84VKK235 pKa = 10.25AGRR238 pKa = 11.84VGWVQWIHH246 pKa = 5.83PSLRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84VSEE255 pKa = 4.39LEE257 pKa = 3.83
Molecular weight: 27.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14149570 |
101 |
16927 |
495.3 |
55.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.711 ± 0.012 | 1.976 ± 0.013 |
5.228 ± 0.01 | 5.993 ± 0.018 |
4.619 ± 0.011 | 6.133 ± 0.017 |
2.503 ± 0.009 | 5.977 ± 0.013 |
6.129 ± 0.018 | 9.17 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.269 ± 0.006 | 4.955 ± 0.011 |
5.338 ± 0.016 | 3.837 ± 0.012 |
5.071 ± 0.011 | 8.33 ± 0.018 |
6.132 ± 0.02 | 6.257 ± 0.012 |
1.287 ± 0.005 | 3.086 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
