
Cactus mild mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Tobamovirus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
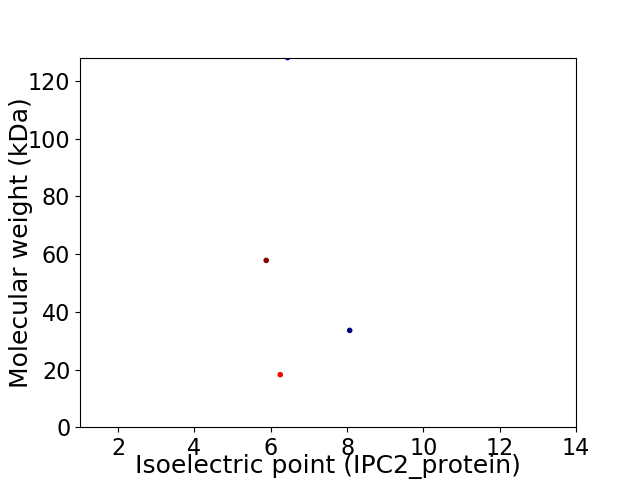
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B8PRG8|B8PRG8_9VIRU Movement protein OS=Cactus mild mottle virus OX=229030 GN=MP PE=4 SV=1
QQ1 pKa = 7.26LTTQPITRR9 pKa = 11.84GVNMYY14 pKa = 10.3IPTAKK19 pKa = 10.54CGDD22 pKa = 3.78PSDD25 pKa = 4.39LQFFYY30 pKa = 11.37DD31 pKa = 4.53AVLPNNSTVLNDD43 pKa = 3.5FDD45 pKa = 6.09SFTLTALPMQLHH57 pKa = 6.23VGDD60 pKa = 4.54CTLDD64 pKa = 3.55LSKK67 pKa = 11.13ADD69 pKa = 3.7PNLYY73 pKa = 10.11ARR75 pKa = 11.84KK76 pKa = 7.94TGFLKK81 pKa = 10.59PVLRR85 pKa = 11.84TGTEE89 pKa = 4.12LPRR92 pKa = 11.84QPNLWEE98 pKa = 4.31NLLALIKK105 pKa = 10.72RR106 pKa = 11.84NFNAPYY112 pKa = 11.11LMGTVDD118 pKa = 3.98IDD120 pKa = 3.66RR121 pKa = 11.84MAEE124 pKa = 4.04SVVNRR129 pKa = 11.84FFEE132 pKa = 4.44VYY134 pKa = 9.42IDD136 pKa = 4.97GSIEE140 pKa = 3.98SCKK143 pKa = 10.25FRR145 pKa = 11.84LLAQNTFAEE154 pKa = 4.42WLKK157 pKa = 10.73LQNPDD162 pKa = 4.81VIGKK166 pKa = 8.76LANFDD171 pKa = 4.67YY172 pKa = 10.88IDD174 pKa = 4.7LPAIDD179 pKa = 4.77QYY181 pKa = 11.93YY182 pKa = 11.11HH183 pKa = 5.92MIKK186 pKa = 10.26RR187 pKa = 11.84KK188 pKa = 9.38PKK190 pKa = 9.61IKK192 pKa = 10.16LDD194 pKa = 3.49QSICTEE200 pKa = 4.1YY201 pKa = 10.11PALQTIVYY209 pKa = 9.09HH210 pKa = 5.82SKK212 pKa = 10.4EE213 pKa = 3.67INAIFGPIFKK223 pKa = 10.34EE224 pKa = 3.79LTRR227 pKa = 11.84VFLDD231 pKa = 3.46SVDD234 pKa = 3.38SSRR237 pKa = 11.84FLFYY241 pKa = 10.18TRR243 pKa = 11.84KK244 pKa = 9.99SADD247 pKa = 3.46DD248 pKa = 3.5VSEE251 pKa = 4.29FFSDD255 pKa = 4.32LPHH258 pKa = 6.81VSQTDD263 pKa = 3.83VYY265 pKa = 10.62EE266 pKa = 4.36LDD268 pKa = 2.8ISKK271 pKa = 10.62YY272 pKa = 10.5DD273 pKa = 3.29KK274 pKa = 11.02SQNEE278 pKa = 3.73LHH280 pKa = 6.64CAIEE284 pKa = 3.96YY285 pKa = 10.08KK286 pKa = 9.93IWEE289 pKa = 4.19RR290 pKa = 11.84LGFDD294 pKa = 3.38GFLRR298 pKa = 11.84DD299 pKa = 3.27VWAQGHH305 pKa = 6.04RR306 pKa = 11.84RR307 pKa = 11.84TVLRR311 pKa = 11.84DD312 pKa = 3.53FQAGIKK318 pKa = 9.98ALIWFQRR325 pKa = 11.84KK326 pKa = 9.56SGDD329 pKa = 3.21VTTFIGNTFINAAAIATLMPLEE351 pKa = 4.43HH352 pKa = 7.02CIKK355 pKa = 10.79AGFCGDD361 pKa = 3.96DD362 pKa = 3.44SVIYY366 pKa = 9.88LPKK369 pKa = 9.05GTPVGDD375 pKa = 3.83VQAKK379 pKa = 10.63ANLEE383 pKa = 4.18WNFEE387 pKa = 3.88AKK389 pKa = 10.2LYY391 pKa = 10.49KK392 pKa = 9.16KK393 pKa = 9.17TYY395 pKa = 10.33GYY397 pKa = 10.63FCGRR401 pKa = 11.84FIIPHH406 pKa = 4.6STGAIVYY413 pKa = 9.27PDD415 pKa = 3.79VLKK418 pKa = 11.33VIAKK422 pKa = 10.34LGAKK426 pKa = 9.33DD427 pKa = 4.07VKK429 pKa = 11.1DD430 pKa = 3.99FDD432 pKa = 3.92HH433 pKa = 8.01LEE435 pKa = 3.95EE436 pKa = 5.1LRR438 pKa = 11.84VSMCDD443 pKa = 3.01TYY445 pKa = 11.47KK446 pKa = 10.81QLGNCAYY453 pKa = 11.0VDD455 pKa = 4.03LLYY458 pKa = 11.17SAMTEE463 pKa = 4.14VYY465 pKa = 9.87PSVVDD470 pKa = 3.53PRR472 pKa = 11.84FAINTVWKK480 pKa = 10.07YY481 pKa = 9.52LTDD484 pKa = 3.19KK485 pKa = 11.04RR486 pKa = 11.84LFKK489 pKa = 10.65SCHH492 pKa = 4.77GFCRR496 pKa = 11.84RR497 pKa = 11.84GPCEE501 pKa = 3.87ALEE504 pKa = 4.36SEE506 pKa = 4.58
QQ1 pKa = 7.26LTTQPITRR9 pKa = 11.84GVNMYY14 pKa = 10.3IPTAKK19 pKa = 10.54CGDD22 pKa = 3.78PSDD25 pKa = 4.39LQFFYY30 pKa = 11.37DD31 pKa = 4.53AVLPNNSTVLNDD43 pKa = 3.5FDD45 pKa = 6.09SFTLTALPMQLHH57 pKa = 6.23VGDD60 pKa = 4.54CTLDD64 pKa = 3.55LSKK67 pKa = 11.13ADD69 pKa = 3.7PNLYY73 pKa = 10.11ARR75 pKa = 11.84KK76 pKa = 7.94TGFLKK81 pKa = 10.59PVLRR85 pKa = 11.84TGTEE89 pKa = 4.12LPRR92 pKa = 11.84QPNLWEE98 pKa = 4.31NLLALIKK105 pKa = 10.72RR106 pKa = 11.84NFNAPYY112 pKa = 11.11LMGTVDD118 pKa = 3.98IDD120 pKa = 3.66RR121 pKa = 11.84MAEE124 pKa = 4.04SVVNRR129 pKa = 11.84FFEE132 pKa = 4.44VYY134 pKa = 9.42IDD136 pKa = 4.97GSIEE140 pKa = 3.98SCKK143 pKa = 10.25FRR145 pKa = 11.84LLAQNTFAEE154 pKa = 4.42WLKK157 pKa = 10.73LQNPDD162 pKa = 4.81VIGKK166 pKa = 8.76LANFDD171 pKa = 4.67YY172 pKa = 10.88IDD174 pKa = 4.7LPAIDD179 pKa = 4.77QYY181 pKa = 11.93YY182 pKa = 11.11HH183 pKa = 5.92MIKK186 pKa = 10.26RR187 pKa = 11.84KK188 pKa = 9.38PKK190 pKa = 9.61IKK192 pKa = 10.16LDD194 pKa = 3.49QSICTEE200 pKa = 4.1YY201 pKa = 10.11PALQTIVYY209 pKa = 9.09HH210 pKa = 5.82SKK212 pKa = 10.4EE213 pKa = 3.67INAIFGPIFKK223 pKa = 10.34EE224 pKa = 3.79LTRR227 pKa = 11.84VFLDD231 pKa = 3.46SVDD234 pKa = 3.38SSRR237 pKa = 11.84FLFYY241 pKa = 10.18TRR243 pKa = 11.84KK244 pKa = 9.99SADD247 pKa = 3.46DD248 pKa = 3.5VSEE251 pKa = 4.29FFSDD255 pKa = 4.32LPHH258 pKa = 6.81VSQTDD263 pKa = 3.83VYY265 pKa = 10.62EE266 pKa = 4.36LDD268 pKa = 2.8ISKK271 pKa = 10.62YY272 pKa = 10.5DD273 pKa = 3.29KK274 pKa = 11.02SQNEE278 pKa = 3.73LHH280 pKa = 6.64CAIEE284 pKa = 3.96YY285 pKa = 10.08KK286 pKa = 9.93IWEE289 pKa = 4.19RR290 pKa = 11.84LGFDD294 pKa = 3.38GFLRR298 pKa = 11.84DD299 pKa = 3.27VWAQGHH305 pKa = 6.04RR306 pKa = 11.84RR307 pKa = 11.84TVLRR311 pKa = 11.84DD312 pKa = 3.53FQAGIKK318 pKa = 9.98ALIWFQRR325 pKa = 11.84KK326 pKa = 9.56SGDD329 pKa = 3.21VTTFIGNTFINAAAIATLMPLEE351 pKa = 4.43HH352 pKa = 7.02CIKK355 pKa = 10.79AGFCGDD361 pKa = 3.96DD362 pKa = 3.44SVIYY366 pKa = 9.88LPKK369 pKa = 9.05GTPVGDD375 pKa = 3.83VQAKK379 pKa = 10.63ANLEE383 pKa = 4.18WNFEE387 pKa = 3.88AKK389 pKa = 10.2LYY391 pKa = 10.49KK392 pKa = 9.16KK393 pKa = 9.17TYY395 pKa = 10.33GYY397 pKa = 10.63FCGRR401 pKa = 11.84FIIPHH406 pKa = 4.6STGAIVYY413 pKa = 9.27PDD415 pKa = 3.79VLKK418 pKa = 11.33VIAKK422 pKa = 10.34LGAKK426 pKa = 9.33DD427 pKa = 4.07VKK429 pKa = 11.1DD430 pKa = 3.99FDD432 pKa = 3.92HH433 pKa = 8.01LEE435 pKa = 3.95EE436 pKa = 5.1LRR438 pKa = 11.84VSMCDD443 pKa = 3.01TYY445 pKa = 11.47KK446 pKa = 10.81QLGNCAYY453 pKa = 11.0VDD455 pKa = 4.03LLYY458 pKa = 11.17SAMTEE463 pKa = 4.14VYY465 pKa = 9.87PSVVDD470 pKa = 3.53PRR472 pKa = 11.84FAINTVWKK480 pKa = 10.07YY481 pKa = 9.52LTDD484 pKa = 3.19KK485 pKa = 11.04RR486 pKa = 11.84LFKK489 pKa = 10.65SCHH492 pKa = 4.77GFCRR496 pKa = 11.84RR497 pKa = 11.84GPCEE501 pKa = 3.87ALEE504 pKa = 4.36SEE506 pKa = 4.58
Molecular weight: 57.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q70P76|Q70P76_9VIRU Capsid protein OS=Cactus mild mottle virus OX=229030 GN=CP PE=3 SV=1
MM1 pKa = 7.29GFVVEE6 pKa = 5.95DD7 pKa = 3.74PAKK10 pKa = 9.64HH11 pKa = 5.75LKK13 pKa = 10.16VNEE16 pKa = 3.87FLGVSGLEE24 pKa = 3.96RR25 pKa = 11.84FKK27 pKa = 11.39SVLTQPFRR35 pKa = 11.84TVSIVNSDD43 pKa = 3.21IVRR46 pKa = 11.84VASAAPLSVPIDD58 pKa = 3.28ILSSLKK64 pKa = 10.5SSKK67 pKa = 9.07LTYY70 pKa = 9.96KK71 pKa = 10.16YY72 pKa = 10.95VYY74 pKa = 9.98VLAVLISGRR83 pKa = 11.84WHH85 pKa = 6.71ISSASPGSVLLVLYY99 pKa = 10.17DD100 pKa = 4.19RR101 pKa = 11.84RR102 pKa = 11.84LQGHH106 pKa = 6.49RR107 pKa = 11.84SCIYY111 pKa = 8.78GGCMSKK117 pKa = 10.48VSANKK122 pKa = 9.52FQVKK126 pKa = 9.79YY127 pKa = 10.75SVGHH131 pKa = 6.07SLTVNDD137 pKa = 5.4FSRR140 pKa = 11.84NPLSLCVALNGVPCDD155 pKa = 4.79DD156 pKa = 3.14GWEE159 pKa = 4.05PLSIEE164 pKa = 4.26VATLLMFTNYY174 pKa = 9.57ILEE177 pKa = 4.33EE178 pKa = 4.19SLTSNILKK186 pKa = 10.02VPPLSFDD193 pKa = 3.76NNSVCLDD200 pKa = 3.24QDD202 pKa = 3.63IIMSKK207 pKa = 10.24FNSVLSTVAVPRR219 pKa = 11.84NVLKK223 pKa = 10.03CTTNFEE229 pKa = 4.5KK230 pKa = 10.71KK231 pKa = 9.86RR232 pKa = 11.84LRR234 pKa = 11.84KK235 pKa = 9.91GKK237 pKa = 8.59WVGNKK242 pKa = 9.89SEE244 pKa = 4.43FVLKK248 pKa = 10.73GDD250 pKa = 3.96TNVDD254 pKa = 3.09PMRR257 pKa = 11.84YY258 pKa = 9.54NGGFLHH264 pKa = 6.0QRR266 pKa = 11.84EE267 pKa = 4.55AEE269 pKa = 4.44HH270 pKa = 6.68ICVPDD275 pKa = 3.61SVVGRR280 pKa = 11.84TGKK283 pKa = 8.11ITQLLNPGATSHH295 pKa = 6.8LSDD298 pKa = 4.3AASSDD303 pKa = 3.86TTCC306 pKa = 4.77
MM1 pKa = 7.29GFVVEE6 pKa = 5.95DD7 pKa = 3.74PAKK10 pKa = 9.64HH11 pKa = 5.75LKK13 pKa = 10.16VNEE16 pKa = 3.87FLGVSGLEE24 pKa = 3.96RR25 pKa = 11.84FKK27 pKa = 11.39SVLTQPFRR35 pKa = 11.84TVSIVNSDD43 pKa = 3.21IVRR46 pKa = 11.84VASAAPLSVPIDD58 pKa = 3.28ILSSLKK64 pKa = 10.5SSKK67 pKa = 9.07LTYY70 pKa = 9.96KK71 pKa = 10.16YY72 pKa = 10.95VYY74 pKa = 9.98VLAVLISGRR83 pKa = 11.84WHH85 pKa = 6.71ISSASPGSVLLVLYY99 pKa = 10.17DD100 pKa = 4.19RR101 pKa = 11.84RR102 pKa = 11.84LQGHH106 pKa = 6.49RR107 pKa = 11.84SCIYY111 pKa = 8.78GGCMSKK117 pKa = 10.48VSANKK122 pKa = 9.52FQVKK126 pKa = 9.79YY127 pKa = 10.75SVGHH131 pKa = 6.07SLTVNDD137 pKa = 5.4FSRR140 pKa = 11.84NPLSLCVALNGVPCDD155 pKa = 4.79DD156 pKa = 3.14GWEE159 pKa = 4.05PLSIEE164 pKa = 4.26VATLLMFTNYY174 pKa = 9.57ILEE177 pKa = 4.33EE178 pKa = 4.19SLTSNILKK186 pKa = 10.02VPPLSFDD193 pKa = 3.76NNSVCLDD200 pKa = 3.24QDD202 pKa = 3.63IIMSKK207 pKa = 10.24FNSVLSTVAVPRR219 pKa = 11.84NVLKK223 pKa = 10.03CTTNFEE229 pKa = 4.5KK230 pKa = 10.71KK231 pKa = 9.86RR232 pKa = 11.84LRR234 pKa = 11.84KK235 pKa = 9.91GKK237 pKa = 8.59WVGNKK242 pKa = 9.89SEE244 pKa = 4.43FVLKK248 pKa = 10.73GDD250 pKa = 3.96TNVDD254 pKa = 3.09PMRR257 pKa = 11.84YY258 pKa = 9.54NGGFLHH264 pKa = 6.0QRR266 pKa = 11.84EE267 pKa = 4.55AEE269 pKa = 4.44HH270 pKa = 6.68ICVPDD275 pKa = 3.61SVVGRR280 pKa = 11.84TGKK283 pKa = 8.11ITQLLNPGATSHH295 pKa = 6.8LSDD298 pKa = 4.3AASSDD303 pKa = 3.86TTCC306 pKa = 4.77
Molecular weight: 33.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2115 |
161 |
1142 |
528.8 |
59.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.139 ± 0.7 | 2.411 ± 0.086 |
6.714 ± 0.494 | 4.397 ± 0.264 |
5.296 ± 0.433 | 5.106 ± 0.275 |
2.222 ± 0.177 | 5.248 ± 0.296 |
6.099 ± 0.472 | 8.889 ± 0.863 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.749 ± 0.209 | 5.012 ± 0.267 |
4.255 ± 0.203 | 3.073 ± 0.491 |
5.059 ± 0.295 | 6.998 ± 1.143 |
6.761 ± 0.782 | 8.652 ± 0.796 |
1.418 ± 0.141 | 3.499 ± 0.384 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
