
Amanita thiersii Skay4041
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Amanitaceae; Amanita; Amanita thiersii
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
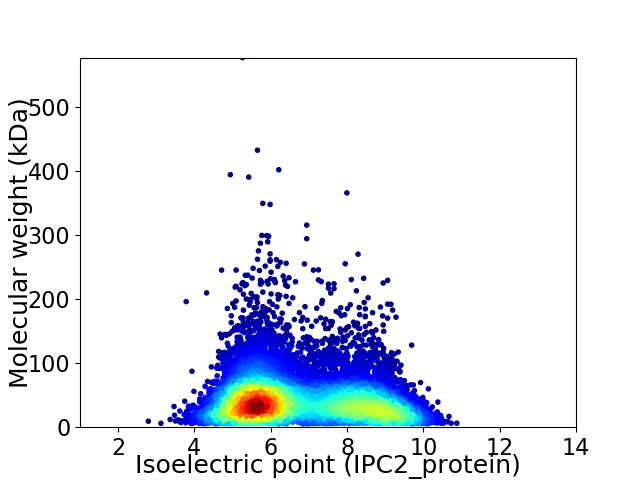
Virtual 2D-PAGE plot for 10343 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A9NXE0|A0A2A9NXE0_9AGAR Uncharacterized protein OS=Amanita thiersii Skay4041 OX=703135 GN=AMATHDRAFT_84058 PE=3 SV=1
MM1 pKa = 7.6ASDD4 pKa = 4.39PLSLTTIARR13 pKa = 11.84NKK15 pKa = 7.45MHH17 pKa = 7.03SSFGSRR23 pKa = 11.84DD24 pKa = 3.27NCSLHH29 pKa = 6.69RR30 pKa = 11.84WVLLKK35 pKa = 11.03NSISVSQSPTLIGATINTLPDD56 pKa = 3.53ASSLCSAGGYY66 pKa = 10.57DD67 pKa = 5.34DD68 pKa = 6.21ADD70 pKa = 3.96DD71 pKa = 4.65RR72 pKa = 11.84NEE74 pKa = 4.44SGDD77 pKa = 3.42SDD79 pKa = 4.69SFFFPDD85 pKa = 3.0AGQIVSSSQTCLSEE99 pKa = 4.5AQWLDD104 pKa = 3.45TLLKK108 pKa = 10.57EE109 pKa = 4.83LGEE112 pKa = 5.14DD113 pKa = 3.6EE114 pKa = 5.56DD115 pKa = 5.6DD116 pKa = 4.96DD117 pKa = 4.66YY118 pKa = 12.07LVEE121 pKa = 5.03SDD123 pKa = 4.18VASPPAPVEE132 pKa = 4.94DD133 pKa = 4.99DD134 pKa = 5.71DD135 pKa = 5.34EE136 pKa = 4.72DD137 pKa = 5.56APLSPLVSPMASSDD151 pKa = 3.75DD152 pKa = 3.86LHH154 pKa = 6.56NQPQYY159 pKa = 11.14YY160 pKa = 9.76SPPRR164 pKa = 11.84QFPYY168 pKa = 10.61PRR170 pKa = 11.84FLHH173 pKa = 6.32PLLHH177 pKa = 6.83PFDD180 pKa = 5.42FDD182 pKa = 3.66QGMQSSISSLPPPYY196 pKa = 10.09DD197 pKa = 4.35DD198 pKa = 4.81PLPYY202 pKa = 10.53HH203 pKa = 6.76NIDD206 pKa = 3.5DD207 pKa = 4.19TQEE210 pKa = 3.93YY211 pKa = 10.01AVPDD215 pKa = 4.31TIIEE219 pKa = 4.87DD220 pKa = 3.62ISDD223 pKa = 4.41DD224 pKa = 4.02EE225 pKa = 5.67SDD227 pKa = 3.82TPSTPSYY234 pKa = 10.19GRR236 pKa = 11.84SSASLSLSDD245 pKa = 4.21IPSIPSPTEE254 pKa = 3.22RR255 pKa = 11.84SSRR258 pKa = 11.84TRR260 pKa = 11.84QATLQVYY267 pKa = 9.93DD268 pKa = 4.32DD269 pKa = 4.3KK270 pKa = 11.65DD271 pKa = 3.78VYY273 pKa = 10.61FYY275 pKa = 10.54PSEE278 pKa = 4.24SDD280 pKa = 3.61PLPFSNDD287 pKa = 2.47HH288 pKa = 6.24HH289 pKa = 7.51SSYY292 pKa = 10.95NAYY295 pKa = 9.74RR296 pKa = 11.84ACC298 pKa = 4.61
MM1 pKa = 7.6ASDD4 pKa = 4.39PLSLTTIARR13 pKa = 11.84NKK15 pKa = 7.45MHH17 pKa = 7.03SSFGSRR23 pKa = 11.84DD24 pKa = 3.27NCSLHH29 pKa = 6.69RR30 pKa = 11.84WVLLKK35 pKa = 11.03NSISVSQSPTLIGATINTLPDD56 pKa = 3.53ASSLCSAGGYY66 pKa = 10.57DD67 pKa = 5.34DD68 pKa = 6.21ADD70 pKa = 3.96DD71 pKa = 4.65RR72 pKa = 11.84NEE74 pKa = 4.44SGDD77 pKa = 3.42SDD79 pKa = 4.69SFFFPDD85 pKa = 3.0AGQIVSSSQTCLSEE99 pKa = 4.5AQWLDD104 pKa = 3.45TLLKK108 pKa = 10.57EE109 pKa = 4.83LGEE112 pKa = 5.14DD113 pKa = 3.6EE114 pKa = 5.56DD115 pKa = 5.6DD116 pKa = 4.96DD117 pKa = 4.66YY118 pKa = 12.07LVEE121 pKa = 5.03SDD123 pKa = 4.18VASPPAPVEE132 pKa = 4.94DD133 pKa = 4.99DD134 pKa = 5.71DD135 pKa = 5.34EE136 pKa = 4.72DD137 pKa = 5.56APLSPLVSPMASSDD151 pKa = 3.75DD152 pKa = 3.86LHH154 pKa = 6.56NQPQYY159 pKa = 11.14YY160 pKa = 9.76SPPRR164 pKa = 11.84QFPYY168 pKa = 10.61PRR170 pKa = 11.84FLHH173 pKa = 6.32PLLHH177 pKa = 6.83PFDD180 pKa = 5.42FDD182 pKa = 3.66QGMQSSISSLPPPYY196 pKa = 10.09DD197 pKa = 4.35DD198 pKa = 4.81PLPYY202 pKa = 10.53HH203 pKa = 6.76NIDD206 pKa = 3.5DD207 pKa = 4.19TQEE210 pKa = 3.93YY211 pKa = 10.01AVPDD215 pKa = 4.31TIIEE219 pKa = 4.87DD220 pKa = 3.62ISDD223 pKa = 4.41DD224 pKa = 4.02EE225 pKa = 5.67SDD227 pKa = 3.82TPSTPSYY234 pKa = 10.19GRR236 pKa = 11.84SSASLSLSDD245 pKa = 4.21IPSIPSPTEE254 pKa = 3.22RR255 pKa = 11.84SSRR258 pKa = 11.84TRR260 pKa = 11.84QATLQVYY267 pKa = 9.93DD268 pKa = 4.32DD269 pKa = 4.3KK270 pKa = 11.65DD271 pKa = 3.78VYY273 pKa = 10.61FYY275 pKa = 10.54PSEE278 pKa = 4.24SDD280 pKa = 3.61PLPFSNDD287 pKa = 2.47HH288 pKa = 6.24HH289 pKa = 7.51SSYY292 pKa = 10.95NAYY295 pKa = 9.74RR296 pKa = 11.84ACC298 pKa = 4.61
Molecular weight: 32.85 kDa
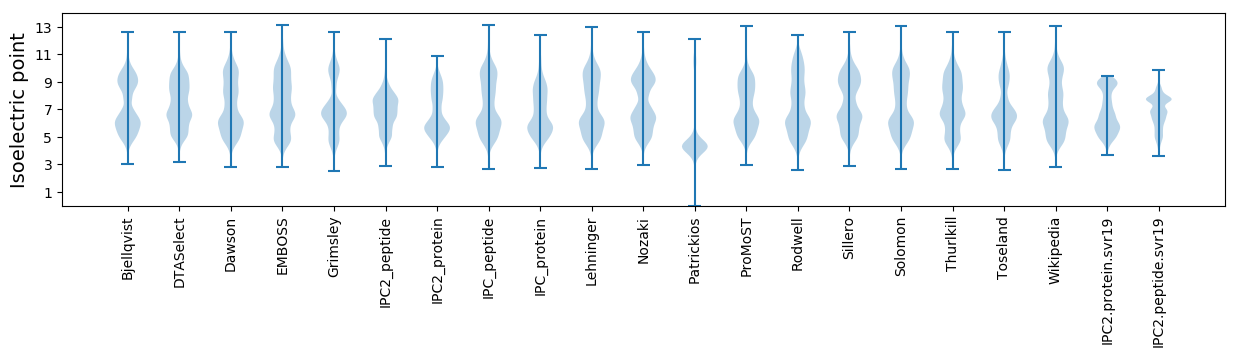
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A9NSE4|A0A2A9NSE4_9AGAR ACT_7 domain-containing protein OS=Amanita thiersii Skay4041 OX=703135 GN=AMATHDRAFT_140371 PE=4 SV=1
SS1 pKa = 6.08HH2 pKa = 7.4TNHH5 pKa = 5.76NQIKK9 pKa = 8.53KK10 pKa = 9.0AHH12 pKa = 6.46RR13 pKa = 11.84NGIKK17 pKa = 9.98KK18 pKa = 8.27PQRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84SLKK28 pKa = 9.88GVRR31 pKa = 11.84FRR33 pKa = 11.84RR34 pKa = 11.84NARR37 pKa = 11.84HH38 pKa = 6.26ALVGSNKK45 pKa = 9.47ARR47 pKa = 11.84SEE49 pKa = 4.14AKK51 pKa = 9.97KK52 pKa = 10.42AAAA55 pKa = 4.29
SS1 pKa = 6.08HH2 pKa = 7.4TNHH5 pKa = 5.76NQIKK9 pKa = 8.53KK10 pKa = 9.0AHH12 pKa = 6.46RR13 pKa = 11.84NGIKK17 pKa = 9.98KK18 pKa = 8.27PQRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84SLKK28 pKa = 9.88GVRR31 pKa = 11.84FRR33 pKa = 11.84RR34 pKa = 11.84NARR37 pKa = 11.84HH38 pKa = 6.26ALVGSNKK45 pKa = 9.47ARR47 pKa = 11.84SEE49 pKa = 4.14AKK51 pKa = 9.97KK52 pKa = 10.42AAAA55 pKa = 4.29
Molecular weight: 6.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4370377 |
49 |
5134 |
422.5 |
46.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.632 ± 0.021 | 1.247 ± 0.009 |
5.393 ± 0.017 | 5.784 ± 0.023 |
3.771 ± 0.016 | 6.404 ± 0.027 |
2.599 ± 0.012 | 5.306 ± 0.017 |
4.814 ± 0.026 | 9.312 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.158 ± 0.009 | 3.87 ± 0.013 |
6.203 ± 0.024 | 4.048 ± 0.017 |
5.951 ± 0.02 | 8.741 ± 0.03 |
6.252 ± 0.015 | 6.273 ± 0.017 |
1.416 ± 0.009 | 2.827 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
