
Gemycircularvirus sp.
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; unclassified Gemycircularvirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
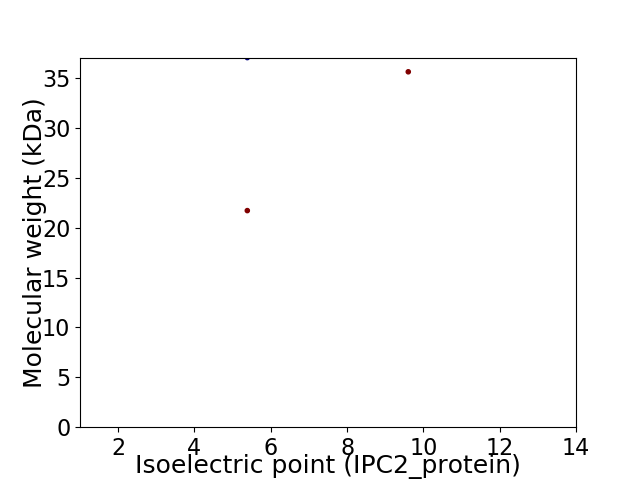
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U3T953|A0A2U3T953_9VIRU Capsid OS=Gemycircularvirus sp. OX=1983771 GN=Cap PE=4 SV=1
MM1 pKa = 7.34TFILNARR8 pKa = 11.84YY9 pKa = 8.98FLVTYY14 pKa = 7.07PQCDD18 pKa = 3.66GLDD21 pKa = 3.2EE22 pKa = 4.42WSVNDD27 pKa = 3.83HH28 pKa = 6.42FGSLGAEE35 pKa = 4.02CIVARR40 pKa = 11.84EE41 pKa = 4.04DD42 pKa = 3.46HH43 pKa = 6.99ADD45 pKa = 3.7GGTHH49 pKa = 6.54LHH51 pKa = 5.76VFCDD55 pKa = 4.88FGRR58 pKa = 11.84KK59 pKa = 8.2FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84ADD66 pKa = 3.04IFDD69 pKa = 3.3VGGFHH74 pKa = 7.62PNIEE78 pKa = 4.01RR79 pKa = 11.84SKK81 pKa = 10.18KK82 pKa = 9.89NPRR85 pKa = 11.84KK86 pKa = 9.57GAAYY90 pKa = 9.66ACKK93 pKa = 10.55DD94 pKa = 3.2GDD96 pKa = 4.08VVAGGLAIPQLPTRR110 pKa = 11.84LVSGAQDD117 pKa = 3.46PGSTLVCAEE126 pKa = 4.1SQRR129 pKa = 11.84EE130 pKa = 4.05FFDD133 pKa = 3.47VAEE136 pKa = 4.29DD137 pKa = 4.97LITKK141 pKa = 9.94FGSFYY146 pKa = 11.46AFARR150 pKa = 11.84WKK152 pKa = 10.03WPEE155 pKa = 3.49LDD157 pKa = 4.28KK158 pKa = 11.31SYY160 pKa = 10.85EE161 pKa = 4.21SPPGLTITDD170 pKa = 3.71GAFPEE175 pKa = 4.23LVRR178 pKa = 11.84YY179 pKa = 7.65RR180 pKa = 11.84TEE182 pKa = 3.78VMGNRR187 pKa = 11.84VNTLCVYY194 pKa = 10.78GEE196 pKa = 4.28SLTGKK201 pKa = 5.81TTWARR206 pKa = 11.84ALGSHH211 pKa = 7.46LYY213 pKa = 9.32MKK215 pKa = 10.74ARR217 pKa = 11.84FNARR221 pKa = 11.84SAMLAEE227 pKa = 4.23GVEE230 pKa = 4.23YY231 pKa = 10.76AVIDD235 pKa = 4.97DD236 pKa = 3.7ISGGIAYY243 pKa = 9.13FPHH246 pKa = 6.21FKK248 pKa = 10.34DD249 pKa = 3.2WFGGQPDD256 pKa = 3.8VQVRR260 pKa = 11.84ALYY263 pKa = 10.51KK264 pKa = 10.64DD265 pKa = 4.06DD266 pKa = 4.41VLLRR270 pKa = 11.84WGKK273 pKa = 7.33PTIWLNQRR281 pKa = 11.84DD282 pKa = 4.25PRR284 pKa = 11.84DD285 pKa = 3.41QLRR288 pKa = 11.84DD289 pKa = 3.57MQGPKK294 pKa = 10.44YY295 pKa = 10.33STDD298 pKa = 3.22QYY300 pKa = 11.19EE301 pKa = 4.84SDD303 pKa = 4.91VKK305 pKa = 10.7WLDD308 pKa = 3.56DD309 pKa = 3.57NCIFVYY315 pKa = 10.26VDD317 pKa = 3.33SPIVTFRR324 pKa = 11.84ASTEE328 pKa = 3.77
MM1 pKa = 7.34TFILNARR8 pKa = 11.84YY9 pKa = 8.98FLVTYY14 pKa = 7.07PQCDD18 pKa = 3.66GLDD21 pKa = 3.2EE22 pKa = 4.42WSVNDD27 pKa = 3.83HH28 pKa = 6.42FGSLGAEE35 pKa = 4.02CIVARR40 pKa = 11.84EE41 pKa = 4.04DD42 pKa = 3.46HH43 pKa = 6.99ADD45 pKa = 3.7GGTHH49 pKa = 6.54LHH51 pKa = 5.76VFCDD55 pKa = 4.88FGRR58 pKa = 11.84KK59 pKa = 8.2FRR61 pKa = 11.84SRR63 pKa = 11.84RR64 pKa = 11.84ADD66 pKa = 3.04IFDD69 pKa = 3.3VGGFHH74 pKa = 7.62PNIEE78 pKa = 4.01RR79 pKa = 11.84SKK81 pKa = 10.18KK82 pKa = 9.89NPRR85 pKa = 11.84KK86 pKa = 9.57GAAYY90 pKa = 9.66ACKK93 pKa = 10.55DD94 pKa = 3.2GDD96 pKa = 4.08VVAGGLAIPQLPTRR110 pKa = 11.84LVSGAQDD117 pKa = 3.46PGSTLVCAEE126 pKa = 4.1SQRR129 pKa = 11.84EE130 pKa = 4.05FFDD133 pKa = 3.47VAEE136 pKa = 4.29DD137 pKa = 4.97LITKK141 pKa = 9.94FGSFYY146 pKa = 11.46AFARR150 pKa = 11.84WKK152 pKa = 10.03WPEE155 pKa = 3.49LDD157 pKa = 4.28KK158 pKa = 11.31SYY160 pKa = 10.85EE161 pKa = 4.21SPPGLTITDD170 pKa = 3.71GAFPEE175 pKa = 4.23LVRR178 pKa = 11.84YY179 pKa = 7.65RR180 pKa = 11.84TEE182 pKa = 3.78VMGNRR187 pKa = 11.84VNTLCVYY194 pKa = 10.78GEE196 pKa = 4.28SLTGKK201 pKa = 5.81TTWARR206 pKa = 11.84ALGSHH211 pKa = 7.46LYY213 pKa = 9.32MKK215 pKa = 10.74ARR217 pKa = 11.84FNARR221 pKa = 11.84SAMLAEE227 pKa = 4.23GVEE230 pKa = 4.23YY231 pKa = 10.76AVIDD235 pKa = 4.97DD236 pKa = 3.7ISGGIAYY243 pKa = 9.13FPHH246 pKa = 6.21FKK248 pKa = 10.34DD249 pKa = 3.2WFGGQPDD256 pKa = 3.8VQVRR260 pKa = 11.84ALYY263 pKa = 10.51KK264 pKa = 10.64DD265 pKa = 4.06DD266 pKa = 4.41VLLRR270 pKa = 11.84WGKK273 pKa = 7.33PTIWLNQRR281 pKa = 11.84DD282 pKa = 4.25PRR284 pKa = 11.84DD285 pKa = 3.41QLRR288 pKa = 11.84DD289 pKa = 3.57MQGPKK294 pKa = 10.44YY295 pKa = 10.33STDD298 pKa = 3.22QYY300 pKa = 11.19EE301 pKa = 4.84SDD303 pKa = 4.91VKK305 pKa = 10.7WLDD308 pKa = 3.56DD309 pKa = 3.57NCIFVYY315 pKa = 10.26VDD317 pKa = 3.33SPIVTFRR324 pKa = 11.84ASTEE328 pKa = 3.77
Molecular weight: 37.05 kDa
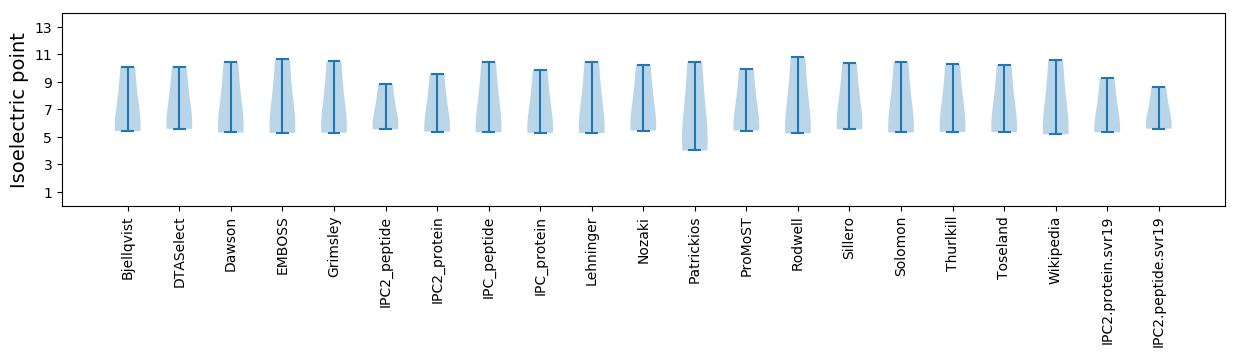
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U3T957|A0A2U3T957_9VIRU RepA OS=Gemycircularvirus sp. OX=1983771 GN=RepA PE=4 SV=1
MM1 pKa = 7.97AYY3 pKa = 9.49RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.53YY7 pKa = 8.93SSYY10 pKa = 9.73RR11 pKa = 11.84ARR13 pKa = 11.84RR14 pKa = 11.84PTKK17 pKa = 10.0RR18 pKa = 11.84YY19 pKa = 5.1GTRR22 pKa = 11.84GAGKK26 pKa = 9.92RR27 pKa = 11.84KK28 pKa = 9.5YY29 pKa = 9.95GSKK32 pKa = 9.82KK33 pKa = 9.39RR34 pKa = 11.84SYY36 pKa = 10.29RR37 pKa = 11.84KK38 pKa = 9.8NPAMSKK44 pKa = 10.29KK45 pKa = 10.31RR46 pKa = 11.84ILNVTSRR53 pKa = 11.84KK54 pKa = 9.6KK55 pKa = 10.5KK56 pKa = 7.97NTMMQYY62 pKa = 11.09VNTDD66 pKa = 3.07STAGTSVSIGPGPMLVSGNSQYY88 pKa = 10.81WGVFTPTAMDD98 pKa = 3.84LTSGADD104 pKa = 3.62LQNSIVDD111 pKa = 3.56SSLRR115 pKa = 11.84TSQTCFIRR123 pKa = 11.84GLRR126 pKa = 11.84EE127 pKa = 3.97NIHH130 pKa = 6.75IEE132 pKa = 4.15TSTGLPWYY140 pKa = 8.09WRR142 pKa = 11.84RR143 pKa = 11.84IVFTAKK149 pKa = 8.65STPWFSFSPNDD160 pKa = 3.76TPTQTNSSSTSAIEE174 pKa = 4.17SNTNGWEE181 pKa = 3.56RR182 pKa = 11.84LYY184 pKa = 11.06FNQYY188 pKa = 9.01VNNAPDD194 pKa = 4.15TISRR198 pKa = 11.84MQRR201 pKa = 11.84DD202 pKa = 4.27LFRR205 pKa = 11.84GEE207 pKa = 4.19QNRR210 pKa = 11.84DD211 pKa = 2.45WSDD214 pKa = 2.85ILTAPIDD221 pKa = 3.68TARR224 pKa = 11.84VDD226 pKa = 3.91LKK228 pKa = 10.92SDD230 pKa = 3.35RR231 pKa = 11.84LRR233 pKa = 11.84VIKK236 pKa = 10.55SGNQVGTNRR245 pKa = 11.84AFKK248 pKa = 10.2MFYY251 pKa = 9.93PCNKK255 pKa = 9.56NLTYY259 pKa = 10.89NDD261 pKa = 4.26DD262 pKa = 3.48EE263 pKa = 5.42SGVVEE268 pKa = 4.21GTTFSSVQDD277 pKa = 3.36KK278 pKa = 11.07RR279 pKa = 11.84GMGNLHH285 pKa = 6.09VVDD288 pKa = 4.62IFLPGTGGAKK298 pKa = 10.25GDD300 pKa = 3.81ILKK303 pKa = 9.03LTSTSTLYY311 pKa = 9.69WHH313 pKa = 7.1EE314 pKa = 4.2KK315 pKa = 9.32
MM1 pKa = 7.97AYY3 pKa = 9.49RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.53YY7 pKa = 8.93SSYY10 pKa = 9.73RR11 pKa = 11.84ARR13 pKa = 11.84RR14 pKa = 11.84PTKK17 pKa = 10.0RR18 pKa = 11.84YY19 pKa = 5.1GTRR22 pKa = 11.84GAGKK26 pKa = 9.92RR27 pKa = 11.84KK28 pKa = 9.5YY29 pKa = 9.95GSKK32 pKa = 9.82KK33 pKa = 9.39RR34 pKa = 11.84SYY36 pKa = 10.29RR37 pKa = 11.84KK38 pKa = 9.8NPAMSKK44 pKa = 10.29KK45 pKa = 10.31RR46 pKa = 11.84ILNVTSRR53 pKa = 11.84KK54 pKa = 9.6KK55 pKa = 10.5KK56 pKa = 7.97NTMMQYY62 pKa = 11.09VNTDD66 pKa = 3.07STAGTSVSIGPGPMLVSGNSQYY88 pKa = 10.81WGVFTPTAMDD98 pKa = 3.84LTSGADD104 pKa = 3.62LQNSIVDD111 pKa = 3.56SSLRR115 pKa = 11.84TSQTCFIRR123 pKa = 11.84GLRR126 pKa = 11.84EE127 pKa = 3.97NIHH130 pKa = 6.75IEE132 pKa = 4.15TSTGLPWYY140 pKa = 8.09WRR142 pKa = 11.84RR143 pKa = 11.84IVFTAKK149 pKa = 8.65STPWFSFSPNDD160 pKa = 3.76TPTQTNSSSTSAIEE174 pKa = 4.17SNTNGWEE181 pKa = 3.56RR182 pKa = 11.84LYY184 pKa = 11.06FNQYY188 pKa = 9.01VNNAPDD194 pKa = 4.15TISRR198 pKa = 11.84MQRR201 pKa = 11.84DD202 pKa = 4.27LFRR205 pKa = 11.84GEE207 pKa = 4.19QNRR210 pKa = 11.84DD211 pKa = 2.45WSDD214 pKa = 2.85ILTAPIDD221 pKa = 3.68TARR224 pKa = 11.84VDD226 pKa = 3.91LKK228 pKa = 10.92SDD230 pKa = 3.35RR231 pKa = 11.84LRR233 pKa = 11.84VIKK236 pKa = 10.55SGNQVGTNRR245 pKa = 11.84AFKK248 pKa = 10.2MFYY251 pKa = 9.93PCNKK255 pKa = 9.56NLTYY259 pKa = 10.89NDD261 pKa = 4.26DD262 pKa = 3.48EE263 pKa = 5.42SGVVEE268 pKa = 4.21GTTFSSVQDD277 pKa = 3.36KK278 pKa = 11.07RR279 pKa = 11.84GMGNLHH285 pKa = 6.09VVDD288 pKa = 4.62IFLPGTGGAKK298 pKa = 10.25GDD300 pKa = 3.81ILKK303 pKa = 9.03LTSTSTLYY311 pKa = 9.69WHH313 pKa = 7.1EE314 pKa = 4.2KK315 pKa = 9.32
Molecular weight: 35.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
835 |
192 |
328 |
278.3 |
31.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.629 ± 1.085 | 1.078 ± 0.503 |
6.467 ± 1.126 | 4.551 ± 1.002 |
4.79 ± 0.696 | 7.186 ± 0.892 |
1.557 ± 0.315 | 3.952 ± 0.269 |
4.91 ± 0.864 | 8.862 ± 2.264 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.874 ± 0.752 | 4.311 ± 1.102 |
5.389 ± 0.863 | 2.874 ± 0.228 |
7.545 ± 0.472 | 8.982 ± 1.649 |
7.665 ± 1.328 | 5.629 ± 0.661 |
2.395 ± 0.083 | 3.353 ± 0.956 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
