
Rat cytomegalovirus (strain Maastricht)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Betaherpesvirinae; Muromegalovirus; Murid betaherpesvirus 2
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
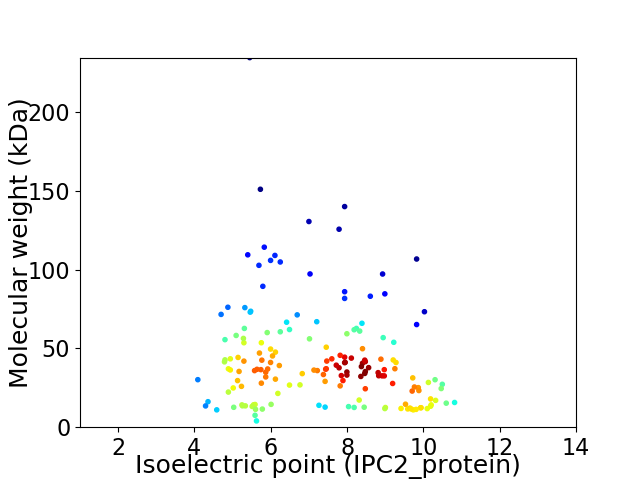
Virtual 2D-PAGE plot for 167 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9DWH6|Q9DWH6_RCMVM Pr3 OS=Rat cytomegalovirus (strain Maastricht) OX=79700 GN=r3 PE=4 SV=1
MM1 pKa = 7.33VPPVDD6 pKa = 3.72TVNGRR11 pKa = 11.84EE12 pKa = 4.05DD13 pKa = 3.16GTLRR17 pKa = 11.84FVFSARR23 pKa = 11.84RR24 pKa = 11.84SGVYY28 pKa = 9.85HH29 pKa = 6.87GLLSLDD35 pKa = 3.55VVKK38 pKa = 10.81KK39 pKa = 8.33FSCVMTVPADD49 pKa = 3.45PCLGARR55 pKa = 11.84QTAAFPSAAAASLPVATTTAEE76 pKa = 3.74NGTRR80 pKa = 11.84PRR82 pKa = 11.84PAGPAPGPPAGGGHH96 pKa = 7.09LDD98 pKa = 3.76VYY100 pKa = 10.97SSTVVFVILSLVLLGGLVMLSVCIWGSSKK129 pKa = 10.93GADD132 pKa = 3.25NGIVLAGDD140 pKa = 3.38SHH142 pKa = 8.41GEE144 pKa = 3.89DD145 pKa = 3.46DD146 pKa = 4.36RR147 pKa = 11.84EE148 pKa = 4.31TVVEE152 pKa = 4.4VVSDD156 pKa = 3.81DD157 pKa = 4.45SEE159 pKa = 5.05VEE161 pKa = 4.22GPDD164 pKa = 3.21VAGDD168 pKa = 3.48VGGDD172 pKa = 3.19RR173 pKa = 11.84RR174 pKa = 11.84DD175 pKa = 3.52DD176 pKa = 4.3DD177 pKa = 4.14GASEE181 pKa = 4.16GKK183 pKa = 10.34EE184 pKa = 3.85EE185 pKa = 4.25AEE187 pKa = 3.99EE188 pKa = 3.99EE189 pKa = 4.27AEE191 pKa = 4.17EE192 pKa = 4.1EE193 pKa = 4.24AEE195 pKa = 4.17EE196 pKa = 3.99EE197 pKa = 4.21AEE199 pKa = 3.96EE200 pKa = 4.26EE201 pKa = 3.97EE202 pKa = 4.42RR203 pKa = 11.84EE204 pKa = 3.95RR205 pKa = 11.84RR206 pKa = 11.84GRR208 pKa = 11.84EE209 pKa = 3.72VEE211 pKa = 3.83PDD213 pKa = 3.7GEE215 pKa = 4.13VDD217 pKa = 2.48PWGRR221 pKa = 11.84FRR223 pKa = 11.84RR224 pKa = 11.84EE225 pKa = 3.6DD226 pKa = 2.98WPYY229 pKa = 10.47PPRR232 pKa = 11.84APPSTSDD239 pKa = 3.28EE240 pKa = 4.24SDD242 pKa = 3.49DD243 pKa = 5.34ARR245 pKa = 11.84DD246 pKa = 3.54SDD248 pKa = 4.33GAAGDD253 pKa = 4.43DD254 pKa = 3.77SSGVCDD260 pKa = 5.91LSTEE264 pKa = 4.17DD265 pKa = 3.93LRR267 pKa = 11.84GTVSVVVTISAVDD280 pKa = 3.54PRR282 pKa = 11.84RR283 pKa = 11.84EE284 pKa = 3.59ADD286 pKa = 3.03AA287 pKa = 5.07
MM1 pKa = 7.33VPPVDD6 pKa = 3.72TVNGRR11 pKa = 11.84EE12 pKa = 4.05DD13 pKa = 3.16GTLRR17 pKa = 11.84FVFSARR23 pKa = 11.84RR24 pKa = 11.84SGVYY28 pKa = 9.85HH29 pKa = 6.87GLLSLDD35 pKa = 3.55VVKK38 pKa = 10.81KK39 pKa = 8.33FSCVMTVPADD49 pKa = 3.45PCLGARR55 pKa = 11.84QTAAFPSAAAASLPVATTTAEE76 pKa = 3.74NGTRR80 pKa = 11.84PRR82 pKa = 11.84PAGPAPGPPAGGGHH96 pKa = 7.09LDD98 pKa = 3.76VYY100 pKa = 10.97SSTVVFVILSLVLLGGLVMLSVCIWGSSKK129 pKa = 10.93GADD132 pKa = 3.25NGIVLAGDD140 pKa = 3.38SHH142 pKa = 8.41GEE144 pKa = 3.89DD145 pKa = 3.46DD146 pKa = 4.36RR147 pKa = 11.84EE148 pKa = 4.31TVVEE152 pKa = 4.4VVSDD156 pKa = 3.81DD157 pKa = 4.45SEE159 pKa = 5.05VEE161 pKa = 4.22GPDD164 pKa = 3.21VAGDD168 pKa = 3.48VGGDD172 pKa = 3.19RR173 pKa = 11.84RR174 pKa = 11.84DD175 pKa = 3.52DD176 pKa = 4.3DD177 pKa = 4.14GASEE181 pKa = 4.16GKK183 pKa = 10.34EE184 pKa = 3.85EE185 pKa = 4.25AEE187 pKa = 3.99EE188 pKa = 3.99EE189 pKa = 4.27AEE191 pKa = 4.17EE192 pKa = 4.1EE193 pKa = 4.24AEE195 pKa = 4.17EE196 pKa = 3.99EE197 pKa = 4.21AEE199 pKa = 3.96EE200 pKa = 4.26EE201 pKa = 3.97EE202 pKa = 4.42RR203 pKa = 11.84EE204 pKa = 3.95RR205 pKa = 11.84RR206 pKa = 11.84GRR208 pKa = 11.84EE209 pKa = 3.72VEE211 pKa = 3.83PDD213 pKa = 3.7GEE215 pKa = 4.13VDD217 pKa = 2.48PWGRR221 pKa = 11.84FRR223 pKa = 11.84RR224 pKa = 11.84EE225 pKa = 3.6DD226 pKa = 2.98WPYY229 pKa = 10.47PPRR232 pKa = 11.84APPSTSDD239 pKa = 3.28EE240 pKa = 4.24SDD242 pKa = 3.49DD243 pKa = 5.34ARR245 pKa = 11.84DD246 pKa = 3.54SDD248 pKa = 4.33GAAGDD253 pKa = 4.43DD254 pKa = 3.77SSGVCDD260 pKa = 5.91LSTEE264 pKa = 4.17DD265 pKa = 3.93LRR267 pKa = 11.84GTVSVVVTISAVDD280 pKa = 3.54PRR282 pKa = 11.84RR283 pKa = 11.84EE284 pKa = 3.59ADD286 pKa = 3.03AA287 pKa = 5.07
Molecular weight: 30.15 kDa
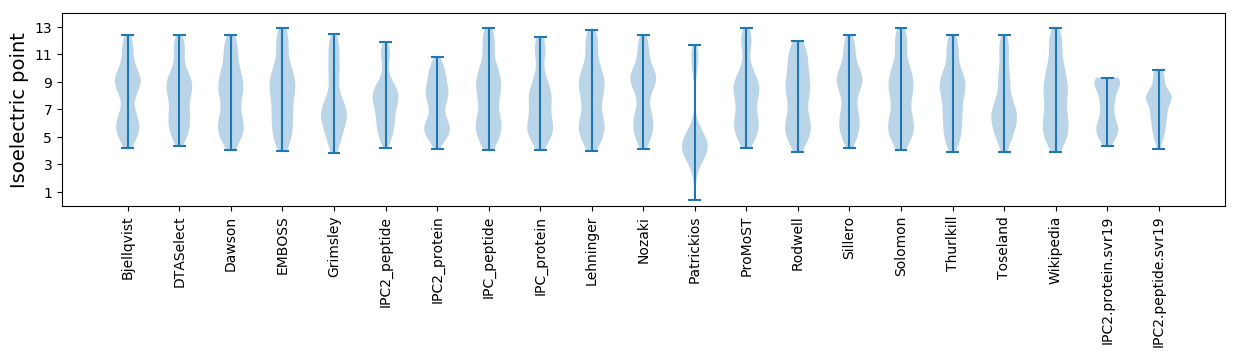
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9DWB3|Q9DWB3_RCMVM PR89 (Fragment) OS=Rat cytomegalovirus (strain Maastricht) OX=79700 GN=R89-EX2 PE=4 SV=1
MM1 pKa = 7.52TFMLAPPMLLLLVCAARR18 pKa = 11.84GRR20 pKa = 11.84GPGGPSPGGAGFALNVTVVKK40 pKa = 11.03GEE42 pKa = 4.25AGPGARR48 pKa = 11.84PAGDD52 pKa = 3.27PTAEE56 pKa = 3.94KK57 pKa = 10.26VRR59 pKa = 11.84RR60 pKa = 11.84AFRR63 pKa = 11.84PVLSLWFRR71 pKa = 11.84AEE73 pKa = 3.93SGRR76 pKa = 11.84KK77 pKa = 7.71FDD79 pKa = 4.09RR80 pKa = 11.84VRR82 pKa = 11.84FLLTCNGTHH91 pKa = 6.55GAVHH95 pKa = 6.89MYY97 pKa = 10.47SPPTTRR103 pKa = 11.84ALSASARR110 pKa = 11.84NGSDD114 pKa = 3.04QLVVQAFSRR123 pKa = 11.84RR124 pKa = 11.84QCAARR129 pKa = 11.84AIRR132 pKa = 11.84LVVTEE137 pKa = 4.67LYY139 pKa = 10.83GPLRR143 pKa = 11.84ALFRR147 pKa = 11.84EE148 pKa = 4.2ACEE151 pKa = 4.26VYY153 pKa = 10.45GASDD157 pKa = 3.9DD158 pKa = 4.57SEE160 pKa = 4.96SSTVTKK166 pKa = 10.8DD167 pKa = 2.88RR168 pKa = 11.84GSSNDD173 pKa = 3.33PYY175 pKa = 11.15ALSSVFVLFMICLAVSFLVCVWALASEE202 pKa = 4.47LKK204 pKa = 10.53KK205 pKa = 10.63RR206 pKa = 11.84ATEE209 pKa = 3.86EE210 pKa = 3.73RR211 pKa = 11.84DD212 pKa = 3.27LEE214 pKa = 4.4GRR216 pKa = 11.84AGTRR220 pKa = 11.84TGGARR225 pKa = 11.84GTTGPAASRR234 pKa = 11.84VRR236 pKa = 11.84EE237 pKa = 4.18TAAVPPPPPTAAEE250 pKa = 4.3SSTSDD255 pKa = 2.88STARR259 pKa = 11.84LLRR262 pKa = 4.41
MM1 pKa = 7.52TFMLAPPMLLLLVCAARR18 pKa = 11.84GRR20 pKa = 11.84GPGGPSPGGAGFALNVTVVKK40 pKa = 11.03GEE42 pKa = 4.25AGPGARR48 pKa = 11.84PAGDD52 pKa = 3.27PTAEE56 pKa = 3.94KK57 pKa = 10.26VRR59 pKa = 11.84RR60 pKa = 11.84AFRR63 pKa = 11.84PVLSLWFRR71 pKa = 11.84AEE73 pKa = 3.93SGRR76 pKa = 11.84KK77 pKa = 7.71FDD79 pKa = 4.09RR80 pKa = 11.84VRR82 pKa = 11.84FLLTCNGTHH91 pKa = 6.55GAVHH95 pKa = 6.89MYY97 pKa = 10.47SPPTTRR103 pKa = 11.84ALSASARR110 pKa = 11.84NGSDD114 pKa = 3.04QLVVQAFSRR123 pKa = 11.84RR124 pKa = 11.84QCAARR129 pKa = 11.84AIRR132 pKa = 11.84LVVTEE137 pKa = 4.67LYY139 pKa = 10.83GPLRR143 pKa = 11.84ALFRR147 pKa = 11.84EE148 pKa = 4.2ACEE151 pKa = 4.26VYY153 pKa = 10.45GASDD157 pKa = 3.9DD158 pKa = 4.57SEE160 pKa = 4.96SSTVTKK166 pKa = 10.8DD167 pKa = 2.88RR168 pKa = 11.84GSSNDD173 pKa = 3.33PYY175 pKa = 11.15ALSSVFVLFMICLAVSFLVCVWALASEE202 pKa = 4.47LKK204 pKa = 10.53KK205 pKa = 10.63RR206 pKa = 11.84ATEE209 pKa = 3.86EE210 pKa = 3.73RR211 pKa = 11.84DD212 pKa = 3.27LEE214 pKa = 4.4GRR216 pKa = 11.84AGTRR220 pKa = 11.84TGGARR225 pKa = 11.84GTTGPAASRR234 pKa = 11.84VRR236 pKa = 11.84EE237 pKa = 4.18TAAVPPPPPTAAEE250 pKa = 4.3SSTSDD255 pKa = 2.88STARR259 pKa = 11.84LLRR262 pKa = 4.41
Molecular weight: 27.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
65855 |
34 |
2100 |
394.3 |
43.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.671 ± 0.328 | 2.089 ± 0.105 |
5.839 ± 0.158 | 5.801 ± 0.17 |
3.787 ± 0.129 | 7.635 ± 0.302 |
2.284 ± 0.103 | 3.833 ± 0.162 |
2.878 ± 0.136 | 8.734 ± 0.226 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.833 ± 0.081 | 3.029 ± 0.153 |
6.335 ± 0.235 | 2.373 ± 0.103 |
9.697 ± 0.297 | 7.509 ± 0.207 |
6.167 ± 0.227 | 7.562 ± 0.204 |
1.072 ± 0.065 | 2.873 ± 0.099 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
