
Human papillomavirus 170
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 7
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
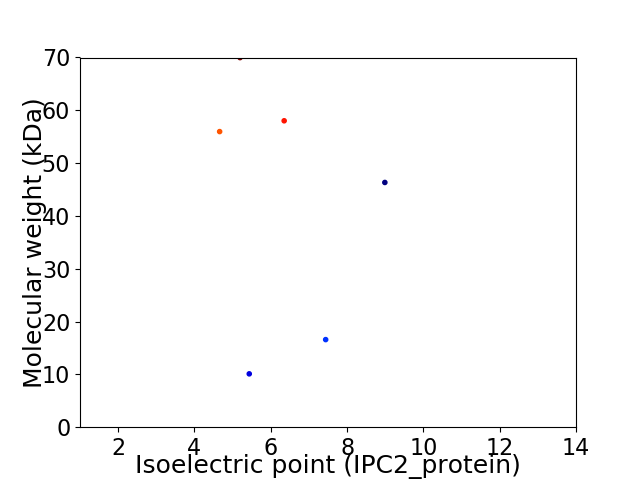
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
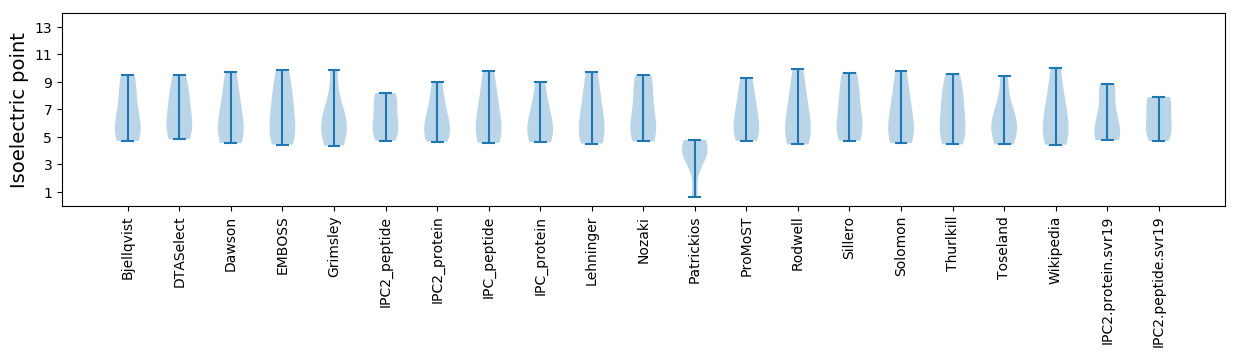
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4MNB7|K4MNB7_9PAPI Protein E7 OS=Human papillomavirus 170 OX=1315265 GN=E7 PE=3 SV=1
MM1 pKa = 7.33EE2 pKa = 4.62SEE4 pKa = 4.43RR5 pKa = 11.84PRR7 pKa = 11.84KK8 pKa = 8.06RR9 pKa = 11.84TKK11 pKa = 10.21RR12 pKa = 11.84DD13 pKa = 3.08SATNLYY19 pKa = 9.71NRR21 pKa = 11.84CQISGNCPEE30 pKa = 4.35DD31 pKa = 3.3VKK33 pKa = 11.62NKK35 pKa = 10.16IEE37 pKa = 4.32GNTLADD43 pKa = 3.78KK44 pKa = 10.6LLKK47 pKa = 9.94ILSSIVYY54 pKa = 9.98FGGLGIGTGRR64 pKa = 11.84GTGGATGYY72 pKa = 10.46RR73 pKa = 11.84PLGSGGGGGGRR84 pKa = 11.84VTPDD88 pKa = 2.82GTVIRR93 pKa = 11.84PNIVVEE99 pKa = 4.05PVGPPEE105 pKa = 4.27IVPIDD110 pKa = 3.79ALSPGSSSIIPMVEE124 pKa = 3.84AGPEE128 pKa = 4.19LIIPEE133 pKa = 4.99TIPDD137 pKa = 3.62LTIGEE142 pKa = 4.54TEE144 pKa = 4.65VITEE148 pKa = 4.69ADD150 pKa = 4.0TIDD153 pKa = 3.12ITGPGGAPAVSTIDD167 pKa = 3.45DD168 pKa = 3.61TSAVIEE174 pKa = 4.3VTPSGPPPRR183 pKa = 11.84RR184 pKa = 11.84VATSTSRR191 pKa = 11.84FSNPTFVSVSTGPTVPQQGATNISVFIDD219 pKa = 3.7GLAGSEE225 pKa = 4.23FVGEE229 pKa = 4.8EE230 pKa = 3.83IPLDD234 pKa = 3.78TFNEE238 pKa = 4.04PQEE241 pKa = 4.4FEE243 pKa = 4.65IEE245 pKa = 5.02DD246 pKa = 3.58IPQPRR251 pKa = 11.84SSTPQAFSRR260 pKa = 11.84AFGRR264 pKa = 11.84ARR266 pKa = 11.84EE267 pKa = 4.14LYY269 pKa = 9.25NRR271 pKa = 11.84RR272 pKa = 11.84VRR274 pKa = 11.84QIRR277 pKa = 11.84TQNINFLTRR286 pKa = 11.84APEE289 pKa = 3.87AVQFTFEE296 pKa = 5.02NPAFDD301 pKa = 4.4NDD303 pKa = 3.25ITLGFQQDD311 pKa = 3.98LDD313 pKa = 3.9QLAAAAPDD321 pKa = 4.13PDD323 pKa = 3.69FADD326 pKa = 2.88IVKK329 pKa = 9.96LHH331 pKa = 6.53RR332 pKa = 11.84PKK334 pKa = 10.62FSEE337 pKa = 4.31TIEE340 pKa = 3.94GRR342 pKa = 11.84VRR344 pKa = 11.84LSRR347 pKa = 11.84LGTKK351 pKa = 8.59GTIRR355 pKa = 11.84LRR357 pKa = 11.84SGTQIGEE364 pKa = 4.53TVHH367 pKa = 7.14LYY369 pKa = 10.96YY370 pKa = 10.71DD371 pKa = 3.42ISSIEE376 pKa = 3.94NADD379 pKa = 4.04AIEE382 pKa = 4.13LSVLGEE388 pKa = 4.03HH389 pKa = 6.72SGDD392 pKa = 3.12ATVINPLAEE401 pKa = 4.28STFVDD406 pKa = 4.02AEE408 pKa = 4.07NSEE411 pKa = 4.29APLLFPEE418 pKa = 4.93QDD420 pKa = 3.67LLDD423 pKa = 5.91DD424 pKa = 3.56ITEE427 pKa = 4.53DD428 pKa = 3.76FSNSHH433 pKa = 6.22IVLSAGGSRR442 pKa = 11.84RR443 pKa = 11.84STLAIPTLPPGVALKK458 pKa = 10.94VFVDD462 pKa = 3.99DD463 pKa = 4.51FGGGLFVSHH472 pKa = 7.73AITTEE477 pKa = 4.44PISNITVPISDD488 pKa = 5.24LGPSILIDD496 pKa = 3.49EE497 pKa = 5.11FSSEE501 pKa = 4.68DD502 pKa = 3.84FVLHH506 pKa = 6.53PSLSKK511 pKa = 9.8KK512 pKa = 9.93RR513 pKa = 11.84KK514 pKa = 8.74RR515 pKa = 11.84KK516 pKa = 9.53RR517 pKa = 11.84LYY519 pKa = 10.76SDD521 pKa = 4.05FF522 pKa = 4.57
MM1 pKa = 7.33EE2 pKa = 4.62SEE4 pKa = 4.43RR5 pKa = 11.84PRR7 pKa = 11.84KK8 pKa = 8.06RR9 pKa = 11.84TKK11 pKa = 10.21RR12 pKa = 11.84DD13 pKa = 3.08SATNLYY19 pKa = 9.71NRR21 pKa = 11.84CQISGNCPEE30 pKa = 4.35DD31 pKa = 3.3VKK33 pKa = 11.62NKK35 pKa = 10.16IEE37 pKa = 4.32GNTLADD43 pKa = 3.78KK44 pKa = 10.6LLKK47 pKa = 9.94ILSSIVYY54 pKa = 9.98FGGLGIGTGRR64 pKa = 11.84GTGGATGYY72 pKa = 10.46RR73 pKa = 11.84PLGSGGGGGGRR84 pKa = 11.84VTPDD88 pKa = 2.82GTVIRR93 pKa = 11.84PNIVVEE99 pKa = 4.05PVGPPEE105 pKa = 4.27IVPIDD110 pKa = 3.79ALSPGSSSIIPMVEE124 pKa = 3.84AGPEE128 pKa = 4.19LIIPEE133 pKa = 4.99TIPDD137 pKa = 3.62LTIGEE142 pKa = 4.54TEE144 pKa = 4.65VITEE148 pKa = 4.69ADD150 pKa = 4.0TIDD153 pKa = 3.12ITGPGGAPAVSTIDD167 pKa = 3.45DD168 pKa = 3.61TSAVIEE174 pKa = 4.3VTPSGPPPRR183 pKa = 11.84RR184 pKa = 11.84VATSTSRR191 pKa = 11.84FSNPTFVSVSTGPTVPQQGATNISVFIDD219 pKa = 3.7GLAGSEE225 pKa = 4.23FVGEE229 pKa = 4.8EE230 pKa = 3.83IPLDD234 pKa = 3.78TFNEE238 pKa = 4.04PQEE241 pKa = 4.4FEE243 pKa = 4.65IEE245 pKa = 5.02DD246 pKa = 3.58IPQPRR251 pKa = 11.84SSTPQAFSRR260 pKa = 11.84AFGRR264 pKa = 11.84ARR266 pKa = 11.84EE267 pKa = 4.14LYY269 pKa = 9.25NRR271 pKa = 11.84RR272 pKa = 11.84VRR274 pKa = 11.84QIRR277 pKa = 11.84TQNINFLTRR286 pKa = 11.84APEE289 pKa = 3.87AVQFTFEE296 pKa = 5.02NPAFDD301 pKa = 4.4NDD303 pKa = 3.25ITLGFQQDD311 pKa = 3.98LDD313 pKa = 3.9QLAAAAPDD321 pKa = 4.13PDD323 pKa = 3.69FADD326 pKa = 2.88IVKK329 pKa = 9.96LHH331 pKa = 6.53RR332 pKa = 11.84PKK334 pKa = 10.62FSEE337 pKa = 4.31TIEE340 pKa = 3.94GRR342 pKa = 11.84VRR344 pKa = 11.84LSRR347 pKa = 11.84LGTKK351 pKa = 8.59GTIRR355 pKa = 11.84LRR357 pKa = 11.84SGTQIGEE364 pKa = 4.53TVHH367 pKa = 7.14LYY369 pKa = 10.96YY370 pKa = 10.71DD371 pKa = 3.42ISSIEE376 pKa = 3.94NADD379 pKa = 4.04AIEE382 pKa = 4.13LSVLGEE388 pKa = 4.03HH389 pKa = 6.72SGDD392 pKa = 3.12ATVINPLAEE401 pKa = 4.28STFVDD406 pKa = 4.02AEE408 pKa = 4.07NSEE411 pKa = 4.29APLLFPEE418 pKa = 4.93QDD420 pKa = 3.67LLDD423 pKa = 5.91DD424 pKa = 3.56ITEE427 pKa = 4.53DD428 pKa = 3.76FSNSHH433 pKa = 6.22IVLSAGGSRR442 pKa = 11.84RR443 pKa = 11.84STLAIPTLPPGVALKK458 pKa = 10.94VFVDD462 pKa = 3.99DD463 pKa = 4.51FGGGLFVSHH472 pKa = 7.73AITTEE477 pKa = 4.44PISNITVPISDD488 pKa = 5.24LGPSILIDD496 pKa = 3.49EE497 pKa = 5.11FSSEE501 pKa = 4.68DD502 pKa = 3.84FVLHH506 pKa = 6.53PSLSKK511 pKa = 9.8KK512 pKa = 9.93RR513 pKa = 11.84KK514 pKa = 8.74RR515 pKa = 11.84KK516 pKa = 9.53RR517 pKa = 11.84LYY519 pKa = 10.76SDD521 pKa = 4.05FF522 pKa = 4.57
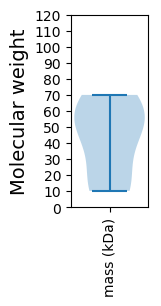
Molecular weight: 55.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4MZ00|K4MZ00_9PAPI Protein E6 OS=Human papillomavirus 170 OX=1315265 GN=E6 PE=3 SV=1
MM1 pKa = 7.71EE2 pKa = 4.81SLEE5 pKa = 4.36DD6 pKa = 4.54RR7 pKa = 11.84FDD9 pKa = 3.98ALQEE13 pKa = 4.05NLLQLYY19 pKa = 8.06EE20 pKa = 4.59AGSDD24 pKa = 3.77NIKK27 pKa = 10.77DD28 pKa = 3.64QILYY32 pKa = 9.31WDD34 pKa = 3.85IVRR37 pKa = 11.84QEE39 pKa = 3.94NVLLHH44 pKa = 5.07YY45 pKa = 10.37ARR47 pKa = 11.84KK48 pKa = 9.91KK49 pKa = 8.74GLNRR53 pKa = 11.84IGLQVIPTLIVSEE66 pKa = 4.49NKK68 pKa = 9.97AKK70 pKa = 10.38QAILMTLQLRR80 pKa = 11.84SLQKK84 pKa = 10.33SAFGSEE90 pKa = 3.77SWTLQDD96 pKa = 3.47TSYY99 pKa = 10.99EE100 pKa = 4.39AYY102 pKa = 9.61TSAPEE107 pKa = 3.89NTFKK111 pKa = 11.04KK112 pKa = 10.57GGFTVDD118 pKa = 2.68VFYY121 pKa = 11.25DD122 pKa = 3.65NDD124 pKa = 3.32EE125 pKa = 4.5DD126 pKa = 4.19NYY128 pKa = 11.19YY129 pKa = 10.18PYY131 pKa = 9.97TAWSYY136 pKa = 11.25IYY138 pKa = 9.65YY139 pKa = 10.53QNGDD143 pKa = 4.31DD144 pKa = 3.39IWYY147 pKa = 9.04KK148 pKa = 10.81VPGQVDD154 pKa = 3.86YY155 pKa = 11.54EE156 pKa = 4.34GLFYY160 pKa = 9.27EE161 pKa = 4.45THH163 pKa = 7.04DD164 pKa = 4.17GEE166 pKa = 4.14KK167 pKa = 10.16QYY169 pKa = 11.63YY170 pKa = 7.34VTFDD174 pKa = 3.0KK175 pKa = 11.17DD176 pKa = 2.86AARR179 pKa = 11.84FSRR182 pKa = 11.84TGMWTVKK189 pKa = 10.32YY190 pKa = 9.68KK191 pKa = 10.88NHH193 pKa = 6.97TISSTSITSTSGHH206 pKa = 7.11PGHH209 pKa = 6.45SPPRR213 pKa = 11.84EE214 pKa = 3.63QSTNSLARR222 pKa = 11.84EE223 pKa = 4.31TEE225 pKa = 3.63ASEE228 pKa = 3.69AGRR231 pKa = 11.84RR232 pKa = 11.84QRR234 pKa = 11.84SSEE237 pKa = 3.84SDD239 pKa = 3.22PRR241 pKa = 11.84RR242 pKa = 11.84SGPLATQSDD251 pKa = 4.66RR252 pKa = 11.84ATSPEE257 pKa = 3.63TSRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84VQRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84GEE271 pKa = 3.69RR272 pKa = 11.84EE273 pKa = 3.33PAAKK277 pKa = 9.87RR278 pKa = 11.84RR279 pKa = 11.84EE280 pKa = 3.76QRR282 pKa = 11.84AARR285 pKa = 11.84GGLSSAPSPEE295 pKa = 4.23EE296 pKa = 3.28VGKK299 pKa = 9.86RR300 pKa = 11.84HH301 pKa = 6.33RR302 pKa = 11.84LVEE305 pKa = 4.4GKK307 pKa = 10.1GLPRR311 pKa = 11.84LRR313 pKa = 11.84RR314 pKa = 11.84LQEE317 pKa = 3.78EE318 pKa = 4.19ARR320 pKa = 11.84DD321 pKa = 3.88PPLVLLKK328 pKa = 11.12GPGNTLKK335 pKa = 10.6CWRR338 pKa = 11.84FKK340 pKa = 10.89CKK342 pKa = 9.83QKK344 pKa = 11.1YY345 pKa = 8.74CGLYY349 pKa = 10.32HH350 pKa = 7.54RR351 pKa = 11.84ISTNFSWVGEE361 pKa = 4.12GSARR365 pKa = 11.84LGTPRR370 pKa = 11.84MLIAFTSPTQRR381 pKa = 11.84QAFLRR386 pKa = 11.84SVVFPKK392 pKa = 10.03GTEE395 pKa = 3.81FSLGNLYY402 pKa = 10.83SLL404 pKa = 4.87
MM1 pKa = 7.71EE2 pKa = 4.81SLEE5 pKa = 4.36DD6 pKa = 4.54RR7 pKa = 11.84FDD9 pKa = 3.98ALQEE13 pKa = 4.05NLLQLYY19 pKa = 8.06EE20 pKa = 4.59AGSDD24 pKa = 3.77NIKK27 pKa = 10.77DD28 pKa = 3.64QILYY32 pKa = 9.31WDD34 pKa = 3.85IVRR37 pKa = 11.84QEE39 pKa = 3.94NVLLHH44 pKa = 5.07YY45 pKa = 10.37ARR47 pKa = 11.84KK48 pKa = 9.91KK49 pKa = 8.74GLNRR53 pKa = 11.84IGLQVIPTLIVSEE66 pKa = 4.49NKK68 pKa = 9.97AKK70 pKa = 10.38QAILMTLQLRR80 pKa = 11.84SLQKK84 pKa = 10.33SAFGSEE90 pKa = 3.77SWTLQDD96 pKa = 3.47TSYY99 pKa = 10.99EE100 pKa = 4.39AYY102 pKa = 9.61TSAPEE107 pKa = 3.89NTFKK111 pKa = 11.04KK112 pKa = 10.57GGFTVDD118 pKa = 2.68VFYY121 pKa = 11.25DD122 pKa = 3.65NDD124 pKa = 3.32EE125 pKa = 4.5DD126 pKa = 4.19NYY128 pKa = 11.19YY129 pKa = 10.18PYY131 pKa = 9.97TAWSYY136 pKa = 11.25IYY138 pKa = 9.65YY139 pKa = 10.53QNGDD143 pKa = 4.31DD144 pKa = 3.39IWYY147 pKa = 9.04KK148 pKa = 10.81VPGQVDD154 pKa = 3.86YY155 pKa = 11.54EE156 pKa = 4.34GLFYY160 pKa = 9.27EE161 pKa = 4.45THH163 pKa = 7.04DD164 pKa = 4.17GEE166 pKa = 4.14KK167 pKa = 10.16QYY169 pKa = 11.63YY170 pKa = 7.34VTFDD174 pKa = 3.0KK175 pKa = 11.17DD176 pKa = 2.86AARR179 pKa = 11.84FSRR182 pKa = 11.84TGMWTVKK189 pKa = 10.32YY190 pKa = 9.68KK191 pKa = 10.88NHH193 pKa = 6.97TISSTSITSTSGHH206 pKa = 7.11PGHH209 pKa = 6.45SPPRR213 pKa = 11.84EE214 pKa = 3.63QSTNSLARR222 pKa = 11.84EE223 pKa = 4.31TEE225 pKa = 3.63ASEE228 pKa = 3.69AGRR231 pKa = 11.84RR232 pKa = 11.84QRR234 pKa = 11.84SSEE237 pKa = 3.84SDD239 pKa = 3.22PRR241 pKa = 11.84RR242 pKa = 11.84SGPLATQSDD251 pKa = 4.66RR252 pKa = 11.84ATSPEE257 pKa = 3.63TSRR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84VQRR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84GEE271 pKa = 3.69RR272 pKa = 11.84EE273 pKa = 3.33PAAKK277 pKa = 9.87RR278 pKa = 11.84RR279 pKa = 11.84EE280 pKa = 3.76QRR282 pKa = 11.84AARR285 pKa = 11.84GGLSSAPSPEE295 pKa = 4.23EE296 pKa = 3.28VGKK299 pKa = 9.86RR300 pKa = 11.84HH301 pKa = 6.33RR302 pKa = 11.84LVEE305 pKa = 4.4GKK307 pKa = 10.1GLPRR311 pKa = 11.84LRR313 pKa = 11.84RR314 pKa = 11.84LQEE317 pKa = 3.78EE318 pKa = 4.19ARR320 pKa = 11.84DD321 pKa = 3.88PPLVLLKK328 pKa = 11.12GPGNTLKK335 pKa = 10.6CWRR338 pKa = 11.84FKK340 pKa = 10.89CKK342 pKa = 9.83QKK344 pKa = 11.1YY345 pKa = 8.74CGLYY349 pKa = 10.32HH350 pKa = 7.54RR351 pKa = 11.84ISTNFSWVGEE361 pKa = 4.12GSARR365 pKa = 11.84LGTPRR370 pKa = 11.84MLIAFTSPTQRR381 pKa = 11.84QAFLRR386 pKa = 11.84SVVFPKK392 pKa = 10.03GTEE395 pKa = 3.81FSLGNLYY402 pKa = 10.83SLL404 pKa = 4.87
Molecular weight: 46.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2300 |
94 |
617 |
383.3 |
42.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.957 ± 0.333 | 2.348 ± 0.942 |
6.0 ± 0.523 | 6.043 ± 0.775 |
4.435 ± 0.275 | 6.522 ± 0.841 |
1.913 ± 0.215 | 5.652 ± 0.801 |
5.13 ± 0.72 | 9.304 ± 0.843 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.304 ± 0.293 | 4.565 ± 0.511 |
5.348 ± 1.003 | 4.217 ± 0.504 |
5.696 ± 1.009 | 8.826 ± 0.23 |
6.435 ± 0.641 | 5.826 ± 0.583 |
1.087 ± 0.315 | 3.391 ± 0.58 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
