
Methylobacterium sp. BTF04
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Methylobacterium; unclassified Methylobacterium
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
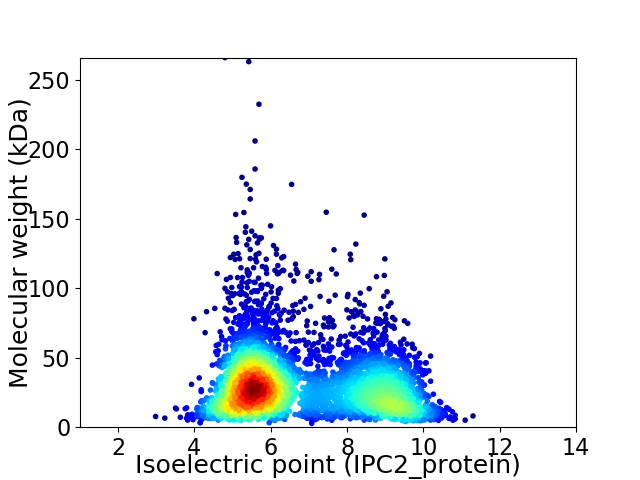
Virtual 2D-PAGE plot for 4688 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M0HUF4|A0A6M0HUF4_9RHIZ Peptide ABC transporter substrate-binding protein OS=Methylobacterium sp. BTF04 OX=2708300 GN=G3T14_18815 PE=3 SV=1
MM1 pKa = 7.85IDD3 pKa = 3.18LTGYY7 pKa = 11.11KK8 pKa = 9.17LTFDD12 pKa = 4.94DD13 pKa = 4.68EE14 pKa = 4.8FNALSVSATGAGTVWGDD31 pKa = 3.45TRR33 pKa = 11.84PGSILRR39 pKa = 11.84PGVDD43 pKa = 2.43IGFGQSAFVDD53 pKa = 4.11PSIGVQPFAVVDD65 pKa = 3.75GAVQIKK71 pKa = 9.56AAPATGATADD81 pKa = 5.03LINPGLWTSGLLQSANSFSQTYY103 pKa = 10.26GYY105 pKa = 11.19FEE107 pKa = 4.25MRR109 pKa = 11.84ADD111 pKa = 3.9LPGTAGTWPGFWLLAANGVWPPEE134 pKa = 3.92LDD136 pKa = 3.09IVEE139 pKa = 4.92AFGADD144 pKa = 3.31PTLTNTVHH152 pKa = 6.54TAQTGTNTYY161 pKa = 7.34QTVYY165 pKa = 8.55THH167 pKa = 6.67EE168 pKa = 4.93PNLLSGFHH176 pKa = 6.15TFGALWTPTTITFTFDD192 pKa = 2.84GFAVGQIATPADD204 pKa = 3.81MHH206 pKa = 6.19SAMYY210 pKa = 9.53PVLALALQRR219 pKa = 11.84DD220 pKa = 4.14VAGITNDD227 pKa = 3.55PKK229 pKa = 10.95TMQVDD234 pKa = 3.55YY235 pKa = 11.32VRR237 pKa = 11.84AYY239 pKa = 10.88SLDD242 pKa = 3.36ASAVEE247 pKa = 4.41VALQTVSSPDD257 pKa = 3.37GVNTANLYY265 pKa = 10.53GAVATNSAPAPVPVAPGADD284 pKa = 3.06TLTIHH289 pKa = 6.87VSGDD293 pKa = 3.04SWNGAPTFILSVDD306 pKa = 3.71GQRR309 pKa = 11.84VGGLFDD315 pKa = 3.48VTALHH320 pKa = 6.84AQGQTQDD327 pKa = 3.65VTVHH331 pKa = 6.46GDD333 pKa = 3.62FGTGPHH339 pKa = 5.47QVSITYY345 pKa = 10.44INDD348 pKa = 3.29ANSNPSYY355 pKa = 10.78DD356 pKa = 3.44AAHH359 pKa = 5.81NVVLNDD365 pKa = 3.2RR366 pKa = 11.84NLFVSGLEE374 pKa = 4.27LDD376 pKa = 3.77GTHH379 pKa = 6.55YY380 pKa = 9.64GTGAVVSNTASVGWDD395 pKa = 3.06KK396 pKa = 11.4LDD398 pKa = 4.02PNAAVMAANGTVVFQTQAAAGPVVDD423 pKa = 4.91PTPPVPTPTPTPTPTPTPTPTPPVTPTTPVAAGSDD458 pKa = 3.46TLTIHH463 pKa = 7.39ISGDD467 pKa = 3.12SWNGAPAFTLAVDD480 pKa = 3.76GHH482 pKa = 5.26QVGGPFDD489 pKa = 3.81VTALHH494 pKa = 6.75AQGQSQDD501 pKa = 3.25ITVHH505 pKa = 6.31GDD507 pKa = 3.32FGTGSHH513 pKa = 5.44QVAITYY519 pKa = 10.8GNDD522 pKa = 3.11ANSNPTYY529 pKa = 10.53DD530 pKa = 3.33AVHH533 pKa = 5.86NVVLNDD539 pKa = 3.34RR540 pKa = 11.84NLFVSGIDD548 pKa = 4.28LDD550 pKa = 4.1GTHH553 pKa = 6.68YY554 pKa = 9.71GTDD557 pKa = 3.07AVFSNTASAGWDD569 pKa = 3.01RR570 pKa = 11.84LDD572 pKa = 3.81FNAAVMAANGTVVFQTQSAGAPVVDD597 pKa = 4.6PTPTVPTPTPTPPSTPTTPVASGSDD622 pKa = 3.12TLTIHH627 pKa = 7.39ISGDD631 pKa = 3.12SWNGAPAFTLAVDD644 pKa = 3.76GHH646 pKa = 5.26QVGGPFDD653 pKa = 3.81VTALHH658 pKa = 6.75AQGQSQDD665 pKa = 3.25ITVHH669 pKa = 6.36GDD671 pKa = 3.47FGTGPHH677 pKa = 5.34QVAIAYY683 pKa = 9.64FNDD686 pKa = 3.5ANSNPSYY693 pKa = 10.81DD694 pKa = 3.3AANNVVLNDD703 pKa = 3.39RR704 pKa = 11.84NLFVSGIDD712 pKa = 3.96LDD714 pKa = 4.2GSHH717 pKa = 6.93YY718 pKa = 9.03GTSAVVSNTASIGWDD733 pKa = 3.05KK734 pKa = 11.18LDD736 pKa = 3.32PHH738 pKa = 6.87AAVMAADD745 pKa = 5.62GILVFQTSGLVV756 pKa = 3.1
MM1 pKa = 7.85IDD3 pKa = 3.18LTGYY7 pKa = 11.11KK8 pKa = 9.17LTFDD12 pKa = 4.94DD13 pKa = 4.68EE14 pKa = 4.8FNALSVSATGAGTVWGDD31 pKa = 3.45TRR33 pKa = 11.84PGSILRR39 pKa = 11.84PGVDD43 pKa = 2.43IGFGQSAFVDD53 pKa = 4.11PSIGVQPFAVVDD65 pKa = 3.75GAVQIKK71 pKa = 9.56AAPATGATADD81 pKa = 5.03LINPGLWTSGLLQSANSFSQTYY103 pKa = 10.26GYY105 pKa = 11.19FEE107 pKa = 4.25MRR109 pKa = 11.84ADD111 pKa = 3.9LPGTAGTWPGFWLLAANGVWPPEE134 pKa = 3.92LDD136 pKa = 3.09IVEE139 pKa = 4.92AFGADD144 pKa = 3.31PTLTNTVHH152 pKa = 6.54TAQTGTNTYY161 pKa = 7.34QTVYY165 pKa = 8.55THH167 pKa = 6.67EE168 pKa = 4.93PNLLSGFHH176 pKa = 6.15TFGALWTPTTITFTFDD192 pKa = 2.84GFAVGQIATPADD204 pKa = 3.81MHH206 pKa = 6.19SAMYY210 pKa = 9.53PVLALALQRR219 pKa = 11.84DD220 pKa = 4.14VAGITNDD227 pKa = 3.55PKK229 pKa = 10.95TMQVDD234 pKa = 3.55YY235 pKa = 11.32VRR237 pKa = 11.84AYY239 pKa = 10.88SLDD242 pKa = 3.36ASAVEE247 pKa = 4.41VALQTVSSPDD257 pKa = 3.37GVNTANLYY265 pKa = 10.53GAVATNSAPAPVPVAPGADD284 pKa = 3.06TLTIHH289 pKa = 6.87VSGDD293 pKa = 3.04SWNGAPTFILSVDD306 pKa = 3.71GQRR309 pKa = 11.84VGGLFDD315 pKa = 3.48VTALHH320 pKa = 6.84AQGQTQDD327 pKa = 3.65VTVHH331 pKa = 6.46GDD333 pKa = 3.62FGTGPHH339 pKa = 5.47QVSITYY345 pKa = 10.44INDD348 pKa = 3.29ANSNPSYY355 pKa = 10.78DD356 pKa = 3.44AAHH359 pKa = 5.81NVVLNDD365 pKa = 3.2RR366 pKa = 11.84NLFVSGLEE374 pKa = 4.27LDD376 pKa = 3.77GTHH379 pKa = 6.55YY380 pKa = 9.64GTGAVVSNTASVGWDD395 pKa = 3.06KK396 pKa = 11.4LDD398 pKa = 4.02PNAAVMAANGTVVFQTQAAAGPVVDD423 pKa = 4.91PTPPVPTPTPTPTPTPTPTPTPPVTPTTPVAAGSDD458 pKa = 3.46TLTIHH463 pKa = 7.39ISGDD467 pKa = 3.12SWNGAPAFTLAVDD480 pKa = 3.76GHH482 pKa = 5.26QVGGPFDD489 pKa = 3.81VTALHH494 pKa = 6.75AQGQSQDD501 pKa = 3.25ITVHH505 pKa = 6.31GDD507 pKa = 3.32FGTGSHH513 pKa = 5.44QVAITYY519 pKa = 10.8GNDD522 pKa = 3.11ANSNPTYY529 pKa = 10.53DD530 pKa = 3.33AVHH533 pKa = 5.86NVVLNDD539 pKa = 3.34RR540 pKa = 11.84NLFVSGIDD548 pKa = 4.28LDD550 pKa = 4.1GTHH553 pKa = 6.68YY554 pKa = 9.71GTDD557 pKa = 3.07AVFSNTASAGWDD569 pKa = 3.01RR570 pKa = 11.84LDD572 pKa = 3.81FNAAVMAANGTVVFQTQSAGAPVVDD597 pKa = 4.6PTPTVPTPTPTPPSTPTTPVASGSDD622 pKa = 3.12TLTIHH627 pKa = 7.39ISGDD631 pKa = 3.12SWNGAPAFTLAVDD644 pKa = 3.76GHH646 pKa = 5.26QVGGPFDD653 pKa = 3.81VTALHH658 pKa = 6.75AQGQSQDD665 pKa = 3.25ITVHH669 pKa = 6.36GDD671 pKa = 3.47FGTGPHH677 pKa = 5.34QVAIAYY683 pKa = 9.64FNDD686 pKa = 3.5ANSNPSYY693 pKa = 10.81DD694 pKa = 3.3AANNVVLNDD703 pKa = 3.39RR704 pKa = 11.84NLFVSGIDD712 pKa = 3.96LDD714 pKa = 4.2GSHH717 pKa = 6.93YY718 pKa = 9.03GTSAVVSNTASIGWDD733 pKa = 3.05KK734 pKa = 11.18LDD736 pKa = 3.32PHH738 pKa = 6.87AAVMAADD745 pKa = 5.62GILVFQTSGLVV756 pKa = 3.1
Molecular weight: 78.0 kDa
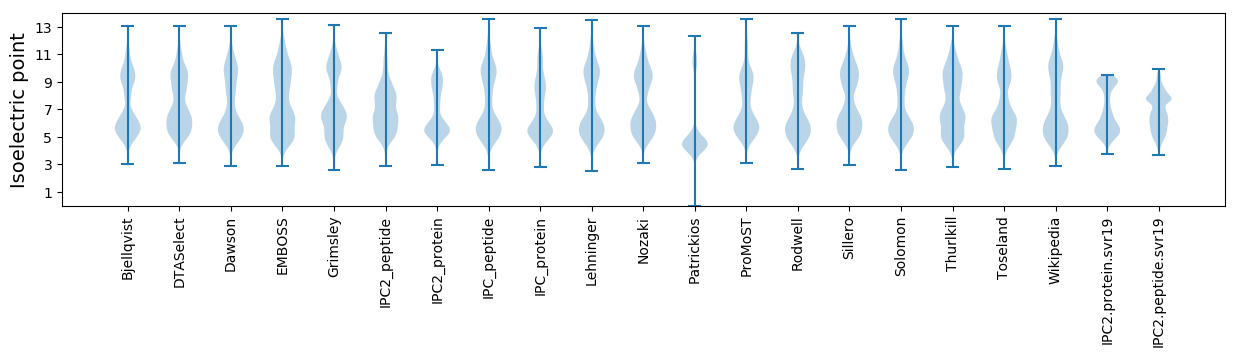
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M0HQH2|A0A6M0HQH2_9RHIZ Porin family protein OS=Methylobacterium sp. BTF04 OX=2708300 GN=G3T14_07645 PE=4 SV=1
MM1 pKa = 7.19HH2 pKa = 7.99RR3 pKa = 11.84IPATALSARR12 pKa = 11.84LLAAAVLSAGLFVFLPSAPARR33 pKa = 11.84AQSLTAPAGQARR45 pKa = 11.84VTAPGGLRR53 pKa = 11.84GRR55 pKa = 11.84AAMRR59 pKa = 11.84HH60 pKa = 4.63HH61 pKa = 6.72RR62 pKa = 11.84QRR64 pKa = 11.84HH65 pKa = 3.8RR66 pKa = 11.84MMHH69 pKa = 5.73HH70 pKa = 6.01RR71 pKa = 11.84RR72 pKa = 11.84HH73 pKa = 5.79RR74 pKa = 3.57
MM1 pKa = 7.19HH2 pKa = 7.99RR3 pKa = 11.84IPATALSARR12 pKa = 11.84LLAAAVLSAGLFVFLPSAPARR33 pKa = 11.84AQSLTAPAGQARR45 pKa = 11.84VTAPGGLRR53 pKa = 11.84GRR55 pKa = 11.84AAMRR59 pKa = 11.84HH60 pKa = 4.63HH61 pKa = 6.72RR62 pKa = 11.84QRR64 pKa = 11.84HH65 pKa = 3.8RR66 pKa = 11.84MMHH69 pKa = 5.73HH70 pKa = 6.01RR71 pKa = 11.84RR72 pKa = 11.84HH73 pKa = 5.79RR74 pKa = 3.57
Molecular weight: 8.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1430968 |
25 |
2510 |
305.2 |
32.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.681 ± 0.057 | 0.822 ± 0.012 |
5.774 ± 0.029 | 5.263 ± 0.033 |
3.442 ± 0.021 | 8.831 ± 0.032 |
2.009 ± 0.018 | 4.826 ± 0.025 |
2.797 ± 0.032 | 10.242 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.16 ± 0.017 | 2.234 ± 0.02 |
5.501 ± 0.032 | 2.822 ± 0.021 |
7.806 ± 0.037 | 5.188 ± 0.025 |
5.684 ± 0.028 | 7.623 ± 0.029 |
1.23 ± 0.013 | 2.064 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
