
Plasmodium falciparum (isolate HB3)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Laverania); Plasmodium falciparum
Average proteome isoelectric point is 7.13
Get precalculated fractions of proteins
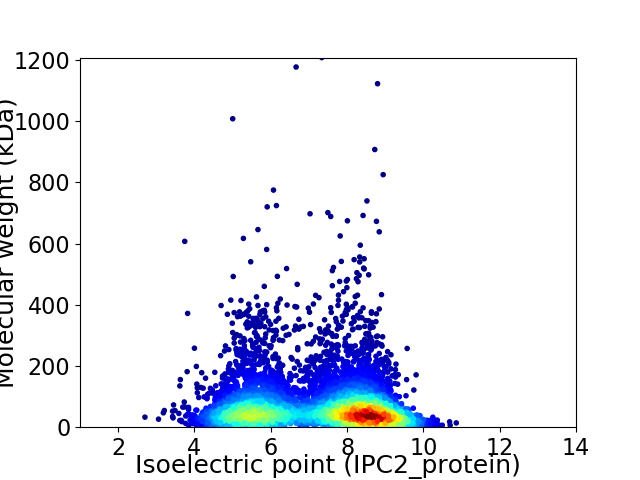
Virtual 2D-PAGE plot for 5452 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0L7K907|A0A0L7K907_PLAFX Uncharacterized protein OS=Plasmodium falciparum (isolate HB3) OX=137071 GN=PFHG_01609 PE=4 SV=1
MM1 pKa = 7.13YY2 pKa = 10.21FIEE5 pKa = 4.9EE6 pKa = 4.1KK7 pKa = 10.16QLYY10 pKa = 9.21YY11 pKa = 10.29PFNEE15 pKa = 3.92TCSRR19 pKa = 11.84YY20 pKa = 9.4RR21 pKa = 11.84RR22 pKa = 11.84IVAEE26 pKa = 4.02EE27 pKa = 3.59QHH29 pKa = 6.61EE30 pKa = 4.31IEE32 pKa = 4.94LDD34 pKa = 3.55EE35 pKa = 4.4EE36 pKa = 4.83SIIVLDD42 pKa = 4.23DD43 pKa = 5.1DD44 pKa = 4.19INIIQEE50 pKa = 4.08EE51 pKa = 4.75YY52 pKa = 11.19EE53 pKa = 4.09NMNSYY58 pKa = 11.21DD59 pKa = 3.87YY60 pKa = 11.86LNLQLYY66 pKa = 10.53NDD68 pKa = 3.85TTMMHH73 pKa = 4.84SHH75 pKa = 6.57EE76 pKa = 4.99VNMQQDD82 pKa = 3.4GDD84 pKa = 3.99INIQQDD90 pKa = 3.17GDD92 pKa = 3.43INMQQDD98 pKa = 3.36GDD100 pKa = 4.26INIQDD105 pKa = 4.33DD106 pKa = 5.15DD107 pKa = 5.05INIQQDD113 pKa = 2.97GDD115 pKa = 3.74INIQQDD121 pKa = 3.07GDD123 pKa = 3.7INIQYY128 pKa = 11.0DD129 pKa = 4.45DD130 pKa = 4.76INIQQDD136 pKa = 2.97GDD138 pKa = 3.83INTEE142 pKa = 3.7QDD144 pKa = 3.25DD145 pKa = 5.38DD146 pKa = 5.12IILLQEE152 pKa = 4.75DD153 pKa = 5.37DD154 pKa = 4.43IDD156 pKa = 4.74LLTDD160 pKa = 3.24EE161 pKa = 6.08DD162 pKa = 4.7INFLEE167 pKa = 4.24QQHH170 pKa = 6.39NDD172 pKa = 3.39LLNEE176 pKa = 3.95EE177 pKa = 4.5SANTNVQNNYY187 pKa = 10.43SFVYY191 pKa = 10.85DD192 pKa = 3.36NDD194 pKa = 3.73YY195 pKa = 11.73NKK197 pKa = 10.36LHH199 pKa = 6.57EE200 pKa = 4.55LAEE203 pKa = 4.4EE204 pKa = 4.36YY205 pKa = 10.81KK206 pKa = 10.3DD207 pKa = 3.57TLNDD211 pKa = 3.41VISRR215 pKa = 11.84MAYY218 pKa = 10.06DD219 pKa = 3.99YY220 pKa = 11.91NDD222 pKa = 3.53LTEE225 pKa = 4.99DD226 pKa = 3.71MNKK229 pKa = 9.46EE230 pKa = 3.38WSFNMWNIRR239 pKa = 11.84WCKK242 pKa = 9.17YY243 pKa = 9.6LEE245 pKa = 3.99NMMEE249 pKa = 4.06EE250 pKa = 3.96VNYY253 pKa = 10.25YY254 pKa = 11.06LNGNFSIDD262 pKa = 3.68DD263 pKa = 3.49KK264 pKa = 11.31KK265 pKa = 11.14QYY267 pKa = 11.14LEE269 pKa = 6.01LLLFWCKK276 pKa = 9.58RR277 pKa = 11.84DD278 pKa = 3.51YY279 pKa = 11.53KK280 pKa = 11.07HH281 pKa = 7.24FIDD284 pKa = 4.4VVKK287 pKa = 10.98KK288 pKa = 9.81EE289 pKa = 3.96WDD291 pKa = 3.37KK292 pKa = 11.63KK293 pKa = 10.97DD294 pKa = 3.66EE295 pKa = 4.49PEE297 pKa = 4.81HH298 pKa = 6.77YY299 pKa = 10.53LEE301 pKa = 4.5RR302 pKa = 5.86
MM1 pKa = 7.13YY2 pKa = 10.21FIEE5 pKa = 4.9EE6 pKa = 4.1KK7 pKa = 10.16QLYY10 pKa = 9.21YY11 pKa = 10.29PFNEE15 pKa = 3.92TCSRR19 pKa = 11.84YY20 pKa = 9.4RR21 pKa = 11.84RR22 pKa = 11.84IVAEE26 pKa = 4.02EE27 pKa = 3.59QHH29 pKa = 6.61EE30 pKa = 4.31IEE32 pKa = 4.94LDD34 pKa = 3.55EE35 pKa = 4.4EE36 pKa = 4.83SIIVLDD42 pKa = 4.23DD43 pKa = 5.1DD44 pKa = 4.19INIIQEE50 pKa = 4.08EE51 pKa = 4.75YY52 pKa = 11.19EE53 pKa = 4.09NMNSYY58 pKa = 11.21DD59 pKa = 3.87YY60 pKa = 11.86LNLQLYY66 pKa = 10.53NDD68 pKa = 3.85TTMMHH73 pKa = 4.84SHH75 pKa = 6.57EE76 pKa = 4.99VNMQQDD82 pKa = 3.4GDD84 pKa = 3.99INIQQDD90 pKa = 3.17GDD92 pKa = 3.43INMQQDD98 pKa = 3.36GDD100 pKa = 4.26INIQDD105 pKa = 4.33DD106 pKa = 5.15DD107 pKa = 5.05INIQQDD113 pKa = 2.97GDD115 pKa = 3.74INIQQDD121 pKa = 3.07GDD123 pKa = 3.7INIQYY128 pKa = 11.0DD129 pKa = 4.45DD130 pKa = 4.76INIQQDD136 pKa = 2.97GDD138 pKa = 3.83INTEE142 pKa = 3.7QDD144 pKa = 3.25DD145 pKa = 5.38DD146 pKa = 5.12IILLQEE152 pKa = 4.75DD153 pKa = 5.37DD154 pKa = 4.43IDD156 pKa = 4.74LLTDD160 pKa = 3.24EE161 pKa = 6.08DD162 pKa = 4.7INFLEE167 pKa = 4.24QQHH170 pKa = 6.39NDD172 pKa = 3.39LLNEE176 pKa = 3.95EE177 pKa = 4.5SANTNVQNNYY187 pKa = 10.43SFVYY191 pKa = 10.85DD192 pKa = 3.36NDD194 pKa = 3.73YY195 pKa = 11.73NKK197 pKa = 10.36LHH199 pKa = 6.57EE200 pKa = 4.55LAEE203 pKa = 4.4EE204 pKa = 4.36YY205 pKa = 10.81KK206 pKa = 10.3DD207 pKa = 3.57TLNDD211 pKa = 3.41VISRR215 pKa = 11.84MAYY218 pKa = 10.06DD219 pKa = 3.99YY220 pKa = 11.91NDD222 pKa = 3.53LTEE225 pKa = 4.99DD226 pKa = 3.71MNKK229 pKa = 9.46EE230 pKa = 3.38WSFNMWNIRR239 pKa = 11.84WCKK242 pKa = 9.17YY243 pKa = 9.6LEE245 pKa = 3.99NMMEE249 pKa = 4.06EE250 pKa = 3.96VNYY253 pKa = 10.25YY254 pKa = 11.06LNGNFSIDD262 pKa = 3.68DD263 pKa = 3.49KK264 pKa = 11.31KK265 pKa = 11.14QYY267 pKa = 11.14LEE269 pKa = 6.01LLLFWCKK276 pKa = 9.58RR277 pKa = 11.84DD278 pKa = 3.51YY279 pKa = 11.53KK280 pKa = 11.07HH281 pKa = 7.24FIDD284 pKa = 4.4VVKK287 pKa = 10.98KK288 pKa = 9.81EE289 pKa = 3.96WDD291 pKa = 3.37KK292 pKa = 11.63KK293 pKa = 10.97DD294 pKa = 3.66EE295 pKa = 4.49PEE297 pKa = 4.81HH298 pKa = 6.77YY299 pKa = 10.53LEE301 pKa = 4.5RR302 pKa = 5.86
Molecular weight: 36.65 kDa
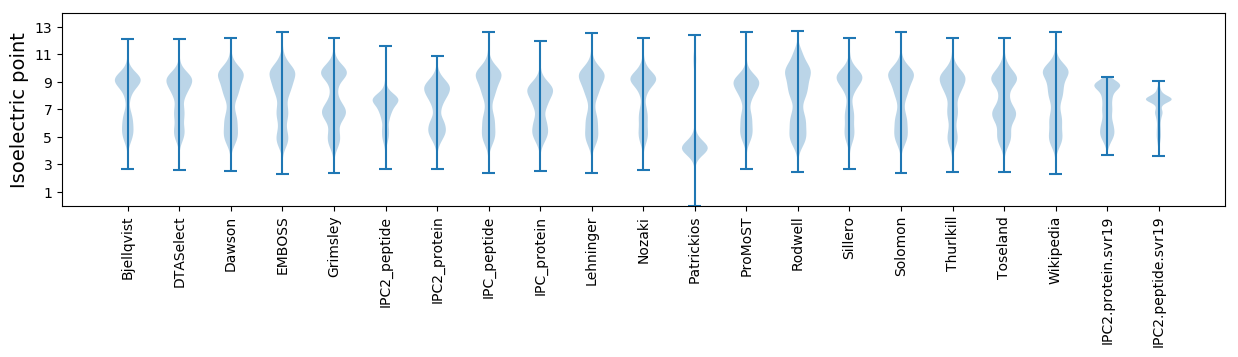
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0L7KD42|A0A0L7KD42_PLAFX Malaria antigen OS=Plasmodium falciparum (isolate HB3) OX=137071 GN=PFHG_03001 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.01KK3 pKa = 10.11DD4 pKa = 3.8RR5 pKa = 11.84EE6 pKa = 4.45PIDD9 pKa = 3.96EE10 pKa = 4.58DD11 pKa = 3.52EE12 pKa = 4.36MRR14 pKa = 11.84ITSTGRR20 pKa = 11.84MTNYY24 pKa = 9.82VNYY27 pKa = 9.53GAKK30 pKa = 9.99ILGDD34 pKa = 3.75EE35 pKa = 4.51DD36 pKa = 3.69KK37 pKa = 11.34KK38 pKa = 11.14SIKK41 pKa = 10.08IKK43 pKa = 9.91ATGNAIGKK51 pKa = 9.51AVTLAEE57 pKa = 4.29IIKK60 pKa = 10.34RR61 pKa = 11.84RR62 pKa = 11.84FKK64 pKa = 10.78GLHH67 pKa = 5.37QITRR71 pKa = 11.84CGSTVITDD79 pKa = 3.53QYY81 pKa = 11.83VSGQDD86 pKa = 3.3NSEE89 pKa = 4.02HH90 pKa = 5.44VVQEE94 pKa = 4.06KK95 pKa = 8.44TVSFIEE101 pKa = 3.91ILLSRR106 pKa = 11.84EE107 pKa = 3.83QLDD110 pKa = 3.78MKK112 pKa = 10.8DD113 pKa = 3.88AGYY116 pKa = 10.2QPPLDD121 pKa = 3.58EE122 pKa = 5.51KK123 pKa = 10.52YY124 pKa = 10.33VKK126 pKa = 10.5EE127 pKa = 4.03MTPEE131 pKa = 4.26EE132 pKa = 4.32IVNSRR137 pKa = 11.84PFRR140 pKa = 11.84RR141 pKa = 11.84GGFRR145 pKa = 11.84PRR147 pKa = 11.84FYY149 pKa = 10.72RR150 pKa = 11.84GFRR153 pKa = 11.84GGRR156 pKa = 11.84GGFLRR161 pKa = 11.84RR162 pKa = 11.84GGYY165 pKa = 9.68RR166 pKa = 11.84GFGDD170 pKa = 3.01RR171 pKa = 11.84VYY173 pKa = 10.69EE174 pKa = 3.93PRR176 pKa = 11.84SSFRR180 pKa = 11.84GGRR183 pKa = 11.84GSGYY187 pKa = 10.08GGNFGRR193 pKa = 11.84GGYY196 pKa = 9.82RR197 pKa = 11.84SGGGMGGGFRR207 pKa = 11.84GGFRR211 pKa = 11.84GGFRR215 pKa = 11.84GGRR218 pKa = 11.84DD219 pKa = 2.89GGYY222 pKa = 9.98RR223 pKa = 11.84GGNRR227 pKa = 11.84GGSRR231 pKa = 11.84SGFRR235 pKa = 11.84GGRR238 pKa = 11.84GGFRR242 pKa = 11.84GGRR245 pKa = 11.84ALSS248 pKa = 3.5
MM1 pKa = 7.44KK2 pKa = 10.01KK3 pKa = 10.11DD4 pKa = 3.8RR5 pKa = 11.84EE6 pKa = 4.45PIDD9 pKa = 3.96EE10 pKa = 4.58DD11 pKa = 3.52EE12 pKa = 4.36MRR14 pKa = 11.84ITSTGRR20 pKa = 11.84MTNYY24 pKa = 9.82VNYY27 pKa = 9.53GAKK30 pKa = 9.99ILGDD34 pKa = 3.75EE35 pKa = 4.51DD36 pKa = 3.69KK37 pKa = 11.34KK38 pKa = 11.14SIKK41 pKa = 10.08IKK43 pKa = 9.91ATGNAIGKK51 pKa = 9.51AVTLAEE57 pKa = 4.29IIKK60 pKa = 10.34RR61 pKa = 11.84RR62 pKa = 11.84FKK64 pKa = 10.78GLHH67 pKa = 5.37QITRR71 pKa = 11.84CGSTVITDD79 pKa = 3.53QYY81 pKa = 11.83VSGQDD86 pKa = 3.3NSEE89 pKa = 4.02HH90 pKa = 5.44VVQEE94 pKa = 4.06KK95 pKa = 8.44TVSFIEE101 pKa = 3.91ILLSRR106 pKa = 11.84EE107 pKa = 3.83QLDD110 pKa = 3.78MKK112 pKa = 10.8DD113 pKa = 3.88AGYY116 pKa = 10.2QPPLDD121 pKa = 3.58EE122 pKa = 5.51KK123 pKa = 10.52YY124 pKa = 10.33VKK126 pKa = 10.5EE127 pKa = 4.03MTPEE131 pKa = 4.26EE132 pKa = 4.32IVNSRR137 pKa = 11.84PFRR140 pKa = 11.84RR141 pKa = 11.84GGFRR145 pKa = 11.84PRR147 pKa = 11.84FYY149 pKa = 10.72RR150 pKa = 11.84GFRR153 pKa = 11.84GGRR156 pKa = 11.84GGFLRR161 pKa = 11.84RR162 pKa = 11.84GGYY165 pKa = 9.68RR166 pKa = 11.84GFGDD170 pKa = 3.01RR171 pKa = 11.84VYY173 pKa = 10.69EE174 pKa = 3.93PRR176 pKa = 11.84SSFRR180 pKa = 11.84GGRR183 pKa = 11.84GSGYY187 pKa = 10.08GGNFGRR193 pKa = 11.84GGYY196 pKa = 9.82RR197 pKa = 11.84SGGGMGGGFRR207 pKa = 11.84GGFRR211 pKa = 11.84GGFRR215 pKa = 11.84GGRR218 pKa = 11.84DD219 pKa = 2.89GGYY222 pKa = 9.98RR223 pKa = 11.84GGNRR227 pKa = 11.84GGSRR231 pKa = 11.84SGFRR235 pKa = 11.84GGRR238 pKa = 11.84GGFRR242 pKa = 11.84GGRR245 pKa = 11.84ALSS248 pKa = 3.5
Molecular weight: 27.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3784517 |
21 |
10299 |
694.2 |
81.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.965 ± 0.023 | 1.771 ± 0.015 |
6.52 ± 0.029 | 7.149 ± 0.061 |
4.327 ± 0.027 | 2.857 ± 0.026 |
2.422 ± 0.014 | 9.193 ± 0.037 |
11.711 ± 0.044 | 7.546 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.199 ± 0.014 | 14.403 ± 0.094 |
1.999 ± 0.02 | 2.777 ± 0.019 |
2.655 ± 0.018 | 6.414 ± 0.025 |
4.102 ± 0.018 | 3.807 ± 0.028 |
0.512 ± 0.009 | 5.671 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
