
Roseivivax sediminis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseivivax
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
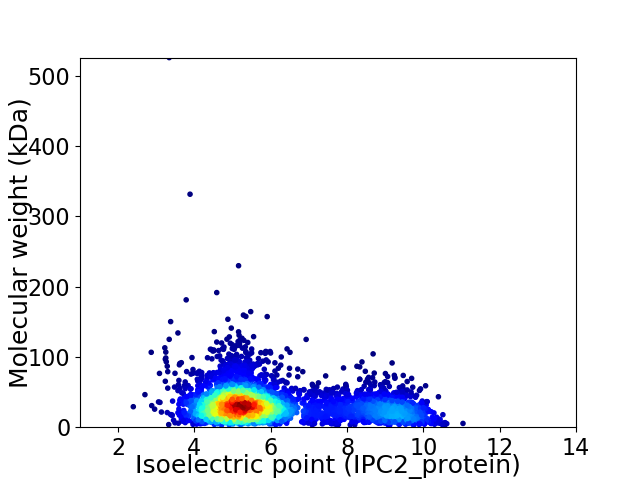
Virtual 2D-PAGE plot for 4371 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I2DWI9|A0A1I2DWI9_9RHOB Alpha-D-ribose 1-methylphosphonate 5-triphosphate diphosphatase OS=Roseivivax sediminis OX=936889 GN=SAMN04515678_11913 PE=4 SV=1
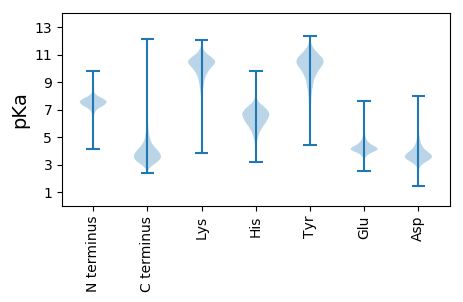
DDD2 pKa = 4.11EE3 pKa = 5.53DD4 pKa = 4.13GYYY7 pKa = 10.01PPNVAPEEE15 pKa = 4.02ADDD18 pKa = 3.76DD19 pKa = 4.3FATPEEE25 pKa = 3.99DD26 pKa = 3.72ALEEE30 pKa = 4.02TAAALLDDD38 pKa = 3.93DDD40 pKa = 4.07DDD42 pKa = 4.72NGDDD46 pKa = 3.87LEEE49 pKa = 4.36LSVTAPAHHH58 pKa = 5.08TLTGTASGWLYYY70 pKa = 10.54PAAGYYY76 pKa = 10.0GPDDD80 pKa = 3.13FTYYY84 pKa = 9.95VGDDD88 pKa = 3.67TATDDD93 pKa = 3.78ATVAVMVGEEE103 pKa = 4.71GAVSDDD109 pKa = 4.76DD110 pKa = 3.84ASGEEE115 pKa = 4.44DDD117 pKa = 3.7AWVFQGIAGSAGLGYYY133 pKa = 10.64GPDDD137 pKa = 3.28LATITSPEEE146 pKa = 4.16VKKK149 pKa = 8.89TAADDD154 pKa = 3.51LTAPRR159 pKa = 11.84LTQAVLDDD167 pKa = 4.25DDD169 pKa = 4.17QISAGFLSEEE179 pKa = 4.66SQQFQEEE186 pKa = 4.33HH187 pKa = 6.29LLVIEEE193 pKa = 6.08DD194 pKa = 3.68EE195 pKa = 4.67NWIRR199 pKa = 11.84FDDD202 pKa = 4.2AYYY205 pKa = 9.96GDDD208 pKa = 4.06LSLIVGTIEEE218 pKa = 4.11GSTTFPLFQTVPSGTVEEE236 pKa = 4.0EE237 pKa = 4.6RR238 pKa = 11.84VTRR241 pKa = 11.84VGDDD245 pKa = 3.32FTFEEE250 pKa = 4.3RR251 pKa = 11.84GGGSSDDD258 pKa = 2.85AQVFSLVRR266 pKa = 11.84AMDDD270 pKa = 3.52TEEE273 pKa = 4.15GVFAGSTSYYY283 pKa = 9.49TDDD286 pKa = 3.1PGYYY290 pKa = 7.24ALVDDD295 pKa = 4.03YY296 pKa = 10.27EEE298 pKa = 5.16AADDD302 pKa = 4.14IGIEEE307 pKa = 4.34DD308 pKa = 4.15SYYY311 pKa = 10.61PQNHHH316 pKa = 6.42PVAVDDD322 pKa = 4.43DD323 pKa = 4.48FSGSGAIVLSKKK335 pKa = 11.14DDD337 pKa = 4.32LANDDD342 pKa = 4.35DDD344 pKa = 4.36DDD346 pKa = 4.32DDD348 pKa = 4.17DD349 pKa = 4.32LSVTLAGPASHHH361 pKa = 6.96SAIDDD366 pKa = 3.75GDDD369 pKa = 3.18TITYYY374 pKa = 9.02PDDD377 pKa = 3.09DDD379 pKa = 4.16EEE381 pKa = 4.48TDDD384 pKa = 3.14FSYYY388 pKa = 10.51LSDDD392 pKa = 3.56TATDDD397 pKa = 3.88EE398 pKa = 3.92WVYYY402 pKa = 11.19DDD404 pKa = 3.48VPVIEEE410 pKa = 4.1WYYY413 pKa = 10.82DDD415 pKa = 3.63QTFGSPGEEE424 pKa = 4.08QTWINILGNVSAEEE438 pKa = 4.16TEEE441 pKa = 5.28SYYY444 pKa = 10.73LNGGAVRR451 pKa = 11.84PLSIGPDDD459 pKa = 3.18RR460 pKa = 11.84RR461 pKa = 11.84LQEEE465 pKa = 4.19GDDD468 pKa = 3.84NVDDD472 pKa = 2.57AFAEEE477 pKa = 4.3DDD479 pKa = 3.38SAADDD484 pKa = 4.19DD485 pKa = 3.92VTLVATLANGTTVSRR500 pKa = 11.84DDD502 pKa = 3.13TVAYYY507 pKa = 9.96EE508 pKa = 5.11DD509 pKa = 4.61EEE511 pKa = 4.29DD512 pKa = 3.23PRR514 pKa = 11.84NYYY517 pKa = 10.66IDDD520 pKa = 3.59SEEE523 pKa = 3.91ADDD526 pKa = 4.12RR527 pKa = 11.84DDD529 pKa = 3.49VQVVDDD535 pKa = 4.16TWAIGPDDD543 pKa = 3.49VRR545 pKa = 11.84PVDDD549 pKa = 3.91GYYY552 pKa = 11.13DD553 pKa = 3.44LLVVGDDD560 pKa = 4.59GWDDD564 pKa = 3.32YYY566 pKa = 11.01LEEE569 pKa = 4.64TVTTHHH575 pKa = 7.23DD576 pKa = 4.72EEE578 pKa = 4.85VDDD581 pKa = 3.61LGRR584 pKa = 11.84DDD586 pKa = 3.08GAFAIGMLWGGHHH599 pKa = 5.48DDD601 pKa = 4.97DD602 pKa = 6.4IPDDD606 pKa = 4.27QPKKK610 pKa = 9.64GWHHH614 pKa = 6.12GAAFFYYY621 pKa = 8.76EEE623 pKa = 3.72LEEE626 pKa = 3.85HHH628 pKa = 6.63YYY630 pKa = 11.26HH631 pKa = 7.65DD632 pKa = 3.54FEEE635 pKa = 4.49YY636 pKa = 10.72GVNNNVSLEEE646 pKa = 3.85EE647 pKa = 4.34DD648 pKa = 2.91KK649 pKa = 10.89YYY651 pKa = 11.18IIVEEE656 pKa = 3.93HHH658 pKa = 4.77VGIYYY663 pKa = 9.48DD664 pKa = 3.29RR665 pKa = 11.84YYY667 pKa = 10.54MKKK670 pKa = 9.29WEEE673 pKa = 4.69EE674 pKa = 4.25TPEEE678 pKa = 3.72TAWSLDDD685 pKa = 3.83IEEE688 pKa = 4.63FSLDDD693 pKa = 3.47EE694 pKa = 4.47PATGGIYYY702 pKa = 10.68NAHHH706 pKa = 6.2YY707 pKa = 10.57YY708 pKa = 10.34DD709 pKa = 3.82SFGDDD714 pKa = 4.04LVSEEE719 pKa = 4.58TGDDD723 pKa = 4.19DD724 pKa = 3.27VSGSDDD730 pKa = 3.13EE731 pKa = 4.83EEE733 pKa = 3.9LIAVTASDDD742 pKa = 3.79TSGLGEEE749 pKa = 3.67DDD751 pKa = 3.74FTGGGGSDDD760 pKa = 3.4FVLGDDD766 pKa = 3.85SRR768 pKa = 11.84AYYY771 pKa = 10.97YY772 pKa = 10.47DD773 pKa = 4.67DD774 pKa = 4.62LAATAGMADDD784 pKa = 3.82YY785 pKa = 11.43FVWDDD790 pKa = 5.2DDD792 pKa = 3.76TEEE795 pKa = 4.94DD796 pKa = 3.73VQLHHH801 pKa = 6.49SAADDD806 pKa = 3.63YY807 pKa = 11.36LTEEE811 pKa = 4.92DD812 pKa = 4.82SGLPPGTAIWRR823 pKa = 11.84LGGAEEE829 pKa = 4.14EEE831 pKa = 4.5VGHHH835 pKa = 6.09HHH837 pKa = 6.29EE838 pKa = 5.62YYY840 pKa = 11.03LSLTAGMFSFTDDD853 pKa = 3.53DD854 pKa = 4.93IA
DDD2 pKa = 4.11EE3 pKa = 5.53DD4 pKa = 4.13GYYY7 pKa = 10.01PPNVAPEEE15 pKa = 4.02ADDD18 pKa = 3.76DD19 pKa = 4.3FATPEEE25 pKa = 3.99DD26 pKa = 3.72ALEEE30 pKa = 4.02TAAALLDDD38 pKa = 3.93DDD40 pKa = 4.07DDD42 pKa = 4.72NGDDD46 pKa = 3.87LEEE49 pKa = 4.36LSVTAPAHHH58 pKa = 5.08TLTGTASGWLYYY70 pKa = 10.54PAAGYYY76 pKa = 10.0GPDDD80 pKa = 3.13FTYYY84 pKa = 9.95VGDDD88 pKa = 3.67TATDDD93 pKa = 3.78ATVAVMVGEEE103 pKa = 4.71GAVSDDD109 pKa = 4.76DD110 pKa = 3.84ASGEEE115 pKa = 4.44DDD117 pKa = 3.7AWVFQGIAGSAGLGYYY133 pKa = 10.64GPDDD137 pKa = 3.28LATITSPEEE146 pKa = 4.16VKKK149 pKa = 8.89TAADDD154 pKa = 3.51LTAPRR159 pKa = 11.84LTQAVLDDD167 pKa = 4.25DDD169 pKa = 4.17QISAGFLSEEE179 pKa = 4.66SQQFQEEE186 pKa = 4.33HH187 pKa = 6.29LLVIEEE193 pKa = 6.08DD194 pKa = 3.68EE195 pKa = 4.67NWIRR199 pKa = 11.84FDDD202 pKa = 4.2AYYY205 pKa = 9.96GDDD208 pKa = 4.06LSLIVGTIEEE218 pKa = 4.11GSTTFPLFQTVPSGTVEEE236 pKa = 4.0EE237 pKa = 4.6RR238 pKa = 11.84VTRR241 pKa = 11.84VGDDD245 pKa = 3.32FTFEEE250 pKa = 4.3RR251 pKa = 11.84GGGSSDDD258 pKa = 2.85AQVFSLVRR266 pKa = 11.84AMDDD270 pKa = 3.52TEEE273 pKa = 4.15GVFAGSTSYYY283 pKa = 9.49TDDD286 pKa = 3.1PGYYY290 pKa = 7.24ALVDDD295 pKa = 4.03YY296 pKa = 10.27EEE298 pKa = 5.16AADDD302 pKa = 4.14IGIEEE307 pKa = 4.34DD308 pKa = 4.15SYYY311 pKa = 10.61PQNHHH316 pKa = 6.42PVAVDDD322 pKa = 4.43DD323 pKa = 4.48FSGSGAIVLSKKK335 pKa = 11.14DDD337 pKa = 4.32LANDDD342 pKa = 4.35DDD344 pKa = 4.36DDD346 pKa = 4.32DDD348 pKa = 4.17DD349 pKa = 4.32LSVTLAGPASHHH361 pKa = 6.96SAIDDD366 pKa = 3.75GDDD369 pKa = 3.18TITYYY374 pKa = 9.02PDDD377 pKa = 3.09DDD379 pKa = 4.16EEE381 pKa = 4.48TDDD384 pKa = 3.14FSYYY388 pKa = 10.51LSDDD392 pKa = 3.56TATDDD397 pKa = 3.88EE398 pKa = 3.92WVYYY402 pKa = 11.19DDD404 pKa = 3.48VPVIEEE410 pKa = 4.1WYYY413 pKa = 10.82DDD415 pKa = 3.63QTFGSPGEEE424 pKa = 4.08QTWINILGNVSAEEE438 pKa = 4.16TEEE441 pKa = 5.28SYYY444 pKa = 10.73LNGGAVRR451 pKa = 11.84PLSIGPDDD459 pKa = 3.18RR460 pKa = 11.84RR461 pKa = 11.84LQEEE465 pKa = 4.19GDDD468 pKa = 3.84NVDDD472 pKa = 2.57AFAEEE477 pKa = 4.3DDD479 pKa = 3.38SAADDD484 pKa = 4.19DD485 pKa = 3.92VTLVATLANGTTVSRR500 pKa = 11.84DDD502 pKa = 3.13TVAYYY507 pKa = 9.96EE508 pKa = 5.11DD509 pKa = 4.61EEE511 pKa = 4.29DD512 pKa = 3.23PRR514 pKa = 11.84NYYY517 pKa = 10.66IDDD520 pKa = 3.59SEEE523 pKa = 3.91ADDD526 pKa = 4.12RR527 pKa = 11.84DDD529 pKa = 3.49VQVVDDD535 pKa = 4.16TWAIGPDDD543 pKa = 3.49VRR545 pKa = 11.84PVDDD549 pKa = 3.91GYYY552 pKa = 11.13DD553 pKa = 3.44LLVVGDDD560 pKa = 4.59GWDDD564 pKa = 3.32YYY566 pKa = 11.01LEEE569 pKa = 4.64TVTTHHH575 pKa = 7.23DD576 pKa = 4.72EEE578 pKa = 4.85VDDD581 pKa = 3.61LGRR584 pKa = 11.84DDD586 pKa = 3.08GAFAIGMLWGGHHH599 pKa = 5.48DDD601 pKa = 4.97DD602 pKa = 6.4IPDDD606 pKa = 4.27QPKKK610 pKa = 9.64GWHHH614 pKa = 6.12GAAFFYYY621 pKa = 8.76EEE623 pKa = 3.72LEEE626 pKa = 3.85HHH628 pKa = 6.63YYY630 pKa = 11.26HH631 pKa = 7.65DD632 pKa = 3.54FEEE635 pKa = 4.49YY636 pKa = 10.72GVNNNVSLEEE646 pKa = 3.85EE647 pKa = 4.34DD648 pKa = 2.91KK649 pKa = 10.89YYY651 pKa = 11.18IIVEEE656 pKa = 3.93HHH658 pKa = 4.77VGIYYY663 pKa = 9.48DD664 pKa = 3.29RR665 pKa = 11.84YYY667 pKa = 10.54MKKK670 pKa = 9.29WEEE673 pKa = 4.69EE674 pKa = 4.25TPEEE678 pKa = 3.72TAWSLDDD685 pKa = 3.83IEEE688 pKa = 4.63FSLDDD693 pKa = 3.47EE694 pKa = 4.47PATGGIYYY702 pKa = 10.68NAHHH706 pKa = 6.2YY707 pKa = 10.57YY708 pKa = 10.34DD709 pKa = 3.82SFGDDD714 pKa = 4.04LVSEEE719 pKa = 4.58TGDDD723 pKa = 4.19DD724 pKa = 3.27VSGSDDD730 pKa = 3.13EE731 pKa = 4.83EEE733 pKa = 3.9LIAVTASDDD742 pKa = 3.79TSGLGEEE749 pKa = 3.67DDD751 pKa = 3.74FTGGGGSDDD760 pKa = 3.4FVLGDDD766 pKa = 3.85SRR768 pKa = 11.84AYYY771 pKa = 10.97YY772 pKa = 10.47DD773 pKa = 4.67DD774 pKa = 4.62LAATAGMADDD784 pKa = 3.82YY785 pKa = 11.43FVWDDD790 pKa = 5.2DDD792 pKa = 3.76TEEE795 pKa = 4.94DD796 pKa = 3.73VQLHHH801 pKa = 6.49SAADDD806 pKa = 3.63YY807 pKa = 11.36LTEEE811 pKa = 4.92DD812 pKa = 4.82SGLPPGTAIWRR823 pKa = 11.84LGGAEEE829 pKa = 4.14EEE831 pKa = 4.5VGHHH835 pKa = 6.09HHH837 pKa = 6.29EE838 pKa = 5.62YYY840 pKa = 11.03LSLTAGMFSFTDDD853 pKa = 3.53DD854 pKa = 4.93IA
Molecular weight: 90.65 kDa
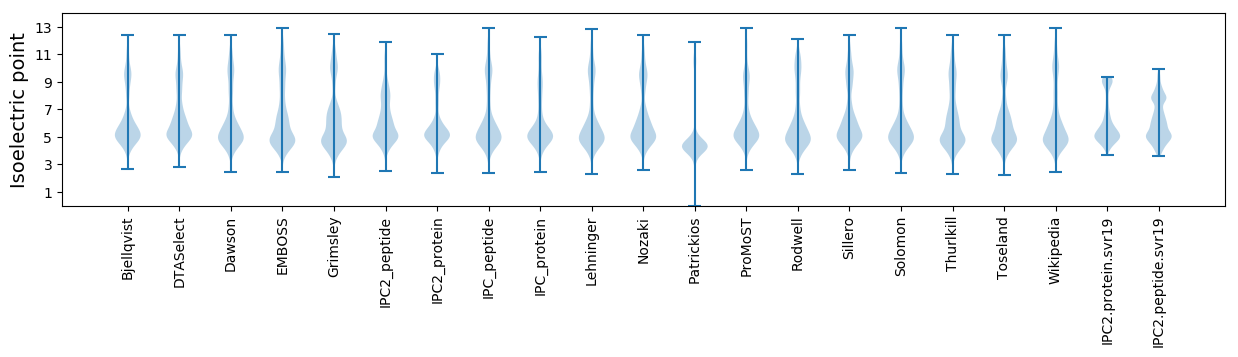
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I2C229|A0A1I2C229_9RHOB DNA-binding transcriptional regulator IclR family OS=Roseivivax sediminis OX=936889 GN=SAMN04515678_11288 PE=4 SV=1
MM1 pKa = 7.84SMLKK5 pKa = 10.42CKK7 pKa = 10.16MPGAALRR14 pKa = 11.84VRR16 pKa = 11.84ASAAALTLSIIAVGASAGAQTLTEE40 pKa = 4.39FTPKK44 pKa = 10.33LPEE47 pKa = 4.07VAVDD51 pKa = 3.62TSPQAVEE58 pKa = 4.02VSLRR62 pKa = 11.84PQMRR66 pKa = 11.84DD67 pKa = 2.79DD68 pKa = 4.46RR69 pKa = 11.84LPKK72 pKa = 10.23ARR74 pKa = 11.84WGDD77 pKa = 3.84SVRR80 pKa = 11.84GVQWTRR86 pKa = 11.84AVVTALRR93 pKa = 11.84GHH95 pKa = 7.19AEE97 pKa = 4.09PLVDD101 pKa = 3.98TVPRR105 pKa = 11.84DD106 pKa = 3.36IDD108 pKa = 3.76DD109 pKa = 3.4WCPAYY114 pKa = 10.33RR115 pKa = 11.84QNNDD119 pKa = 3.13RR120 pKa = 11.84LRR122 pKa = 11.84EE123 pKa = 4.14AFWVGLVSSLAKK135 pKa = 10.39HH136 pKa = 5.73EE137 pKa = 4.67STWRR141 pKa = 11.84PTVVGGGGRR150 pKa = 11.84WHH152 pKa = 7.22GLLQILPSTARR163 pKa = 11.84LYY165 pKa = 11.0GCRR168 pKa = 11.84AQSGSALLSGPPNLSCGLRR187 pKa = 11.84IMAKK191 pKa = 6.56TVRR194 pKa = 11.84RR195 pKa = 11.84DD196 pKa = 3.6GVVSAGMRR204 pKa = 11.84GVAADD209 pKa = 3.73WGPFHH214 pKa = 7.25SRR216 pKa = 11.84TKK218 pKa = 10.11RR219 pKa = 11.84ADD221 pKa = 2.98MMAYY225 pKa = 8.48TRR227 pKa = 11.84NQAFCKK233 pKa = 9.59VVPGTRR239 pKa = 11.84PEE241 pKa = 3.96LRR243 pKa = 11.84PDD245 pKa = 2.8RR246 pKa = 11.84WEE248 pKa = 4.18APVMARR254 pKa = 11.84APHH257 pKa = 6.42HH258 pKa = 6.51PAPDD262 pKa = 3.91GPVLSTQGRR271 pKa = 11.84YY272 pKa = 7.24YY273 pKa = 11.23ARR275 pKa = 4.59
MM1 pKa = 7.84SMLKK5 pKa = 10.42CKK7 pKa = 10.16MPGAALRR14 pKa = 11.84VRR16 pKa = 11.84ASAAALTLSIIAVGASAGAQTLTEE40 pKa = 4.39FTPKK44 pKa = 10.33LPEE47 pKa = 4.07VAVDD51 pKa = 3.62TSPQAVEE58 pKa = 4.02VSLRR62 pKa = 11.84PQMRR66 pKa = 11.84DD67 pKa = 2.79DD68 pKa = 4.46RR69 pKa = 11.84LPKK72 pKa = 10.23ARR74 pKa = 11.84WGDD77 pKa = 3.84SVRR80 pKa = 11.84GVQWTRR86 pKa = 11.84AVVTALRR93 pKa = 11.84GHH95 pKa = 7.19AEE97 pKa = 4.09PLVDD101 pKa = 3.98TVPRR105 pKa = 11.84DD106 pKa = 3.36IDD108 pKa = 3.76DD109 pKa = 3.4WCPAYY114 pKa = 10.33RR115 pKa = 11.84QNNDD119 pKa = 3.13RR120 pKa = 11.84LRR122 pKa = 11.84EE123 pKa = 4.14AFWVGLVSSLAKK135 pKa = 10.39HH136 pKa = 5.73EE137 pKa = 4.67STWRR141 pKa = 11.84PTVVGGGGRR150 pKa = 11.84WHH152 pKa = 7.22GLLQILPSTARR163 pKa = 11.84LYY165 pKa = 11.0GCRR168 pKa = 11.84AQSGSALLSGPPNLSCGLRR187 pKa = 11.84IMAKK191 pKa = 6.56TVRR194 pKa = 11.84RR195 pKa = 11.84DD196 pKa = 3.6GVVSAGMRR204 pKa = 11.84GVAADD209 pKa = 3.73WGPFHH214 pKa = 7.25SRR216 pKa = 11.84TKK218 pKa = 10.11RR219 pKa = 11.84ADD221 pKa = 2.98MMAYY225 pKa = 8.48TRR227 pKa = 11.84NQAFCKK233 pKa = 9.59VVPGTRR239 pKa = 11.84PEE241 pKa = 3.96LRR243 pKa = 11.84PDD245 pKa = 2.8RR246 pKa = 11.84WEE248 pKa = 4.18APVMARR254 pKa = 11.84APHH257 pKa = 6.42HH258 pKa = 6.51PAPDD262 pKa = 3.91GPVLSTQGRR271 pKa = 11.84YY272 pKa = 7.24YY273 pKa = 11.23ARR275 pKa = 4.59
Molecular weight: 29.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1359608 |
24 |
5077 |
311.1 |
33.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.043 ± 0.062 | 0.867 ± 0.013 |
6.257 ± 0.046 | 6.319 ± 0.039 |
3.544 ± 0.025 | 9.109 ± 0.049 |
2.044 ± 0.017 | 4.705 ± 0.029 |
2.445 ± 0.031 | 9.966 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.581 ± 0.02 | 2.224 ± 0.022 |
5.291 ± 0.028 | 2.806 ± 0.024 |
7.449 ± 0.044 | 4.974 ± 0.026 |
5.484 ± 0.03 | 7.368 ± 0.033 |
1.405 ± 0.018 | 2.117 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
