
Testudinid alphaherpesvirus 3
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Scutavirus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
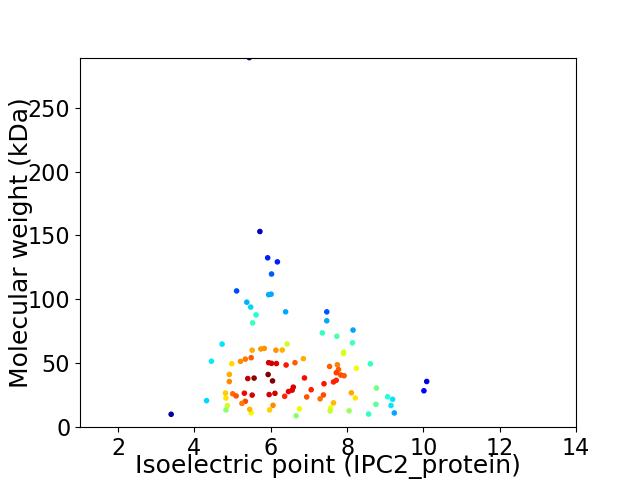
Virtual 2D-PAGE plot for 100 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1R170|A0A0K1R170_9ALPH Envelope glycoprotein L OS=Testudinid alphaherpesvirus 3 OX=2560801 GN=ORF25 PE=4 SV=1
MM1 pKa = 7.69LLTFRR6 pKa = 11.84NPRR9 pKa = 11.84AIRR12 pKa = 11.84DD13 pKa = 3.55IVSIGMFLRR22 pKa = 11.84SVTICGLFLSGFVVCYY38 pKa = 8.67PAQTHH43 pKa = 5.71SPTYY47 pKa = 10.67GGTLPTDD54 pKa = 3.25VSDD57 pKa = 4.17GLRR60 pKa = 11.84STEE63 pKa = 3.76KK64 pKa = 10.07PRR66 pKa = 11.84PAEE69 pKa = 3.74EE70 pKa = 3.71QKK72 pKa = 10.91IIDD75 pKa = 4.23TLNQKK80 pKa = 9.94CVLQHH85 pKa = 6.75DD86 pKa = 4.46VEE88 pKa = 4.82FKK90 pKa = 10.42NCAIQVSFSDD100 pKa = 4.27PEE102 pKa = 3.93ITLALYY108 pKa = 10.52DD109 pKa = 4.38HH110 pKa = 7.12LALYY114 pKa = 7.08QQCVIQFTSFAEE126 pKa = 4.52GTTTCEE132 pKa = 3.61SWGPTPIPLDD142 pKa = 3.52LCLCDD147 pKa = 4.13EE148 pKa = 5.03GGLNKK153 pKa = 10.33CAVSGAKK160 pKa = 9.1TIHH163 pKa = 5.57VFYY166 pKa = 10.11PSGPAVSAAKK176 pKa = 9.63FQKK179 pKa = 10.49NQTLLITTDD188 pKa = 3.17DD189 pKa = 3.87CGRR192 pKa = 11.84TLSTASTASPISDD205 pKa = 3.81SEE207 pKa = 4.2KK208 pKa = 10.3HH209 pKa = 6.57RR210 pKa = 11.84SICFQLSPKK219 pKa = 9.71PEE221 pKa = 4.5NIGQHH226 pKa = 4.82ILRR229 pKa = 11.84VIRR232 pKa = 11.84QPLTAISRR240 pKa = 11.84QGPFSYY246 pKa = 11.14LNDD249 pKa = 3.34THH251 pKa = 6.82TLLLPGPVSQEE262 pKa = 3.52QHH264 pKa = 5.6DD265 pKa = 4.36TYY267 pKa = 11.26DD268 pKa = 3.42GSGEE272 pKa = 4.04MEE274 pKa = 4.6PNWFNYY280 pKa = 8.29TYY282 pKa = 10.94QDD284 pKa = 4.06DD285 pKa = 4.29LTLALYY291 pKa = 10.68SGDD294 pKa = 3.99LLDD297 pKa = 4.8VLGDD301 pKa = 3.52DD302 pKa = 3.54TVFFVFPDD310 pKa = 4.4SICVNYY316 pKa = 9.68TSHH319 pKa = 7.48PYY321 pKa = 10.3SEE323 pKa = 4.72TSAEE327 pKa = 4.13YY328 pKa = 10.73GDD330 pKa = 3.82SSEE333 pKa = 4.05ALEE336 pKa = 4.1YY337 pKa = 10.7TIGRR341 pKa = 11.84VDD343 pKa = 3.54ACTLTAIQTQINGTVIYY360 pKa = 9.81DD361 pKa = 3.62VPCEE365 pKa = 3.9EE366 pKa = 4.55GQTCFGYY373 pKa = 10.87VYY375 pKa = 10.6GDD377 pKa = 3.4RR378 pKa = 11.84MKK380 pKa = 10.59LTVAHH385 pKa = 6.93PGYY388 pKa = 6.98TTQEE392 pKa = 4.09HH393 pKa = 6.71NYY395 pKa = 9.31GNYY398 pKa = 9.6VEE400 pKa = 5.55LDD402 pKa = 3.89AYY404 pKa = 11.22DD405 pKa = 4.3MITPEE410 pKa = 4.52PGSPSKK416 pKa = 10.81HH417 pKa = 6.29IIWKK421 pKa = 5.75TTLFSDD427 pKa = 4.96LSDD430 pKa = 4.35DD431 pKa = 4.88FVDD434 pKa = 3.62TRR436 pKa = 11.84LNRR439 pKa = 11.84HH440 pKa = 5.45QLDD443 pKa = 4.23GVCDD447 pKa = 4.11DD448 pKa = 4.77EE449 pKa = 4.85GCPGEE454 pKa = 4.99GSWEE458 pKa = 4.24DD459 pKa = 3.33IYY461 pKa = 10.75PTVHH465 pKa = 6.33EE466 pKa = 4.45
MM1 pKa = 7.69LLTFRR6 pKa = 11.84NPRR9 pKa = 11.84AIRR12 pKa = 11.84DD13 pKa = 3.55IVSIGMFLRR22 pKa = 11.84SVTICGLFLSGFVVCYY38 pKa = 8.67PAQTHH43 pKa = 5.71SPTYY47 pKa = 10.67GGTLPTDD54 pKa = 3.25VSDD57 pKa = 4.17GLRR60 pKa = 11.84STEE63 pKa = 3.76KK64 pKa = 10.07PRR66 pKa = 11.84PAEE69 pKa = 3.74EE70 pKa = 3.71QKK72 pKa = 10.91IIDD75 pKa = 4.23TLNQKK80 pKa = 9.94CVLQHH85 pKa = 6.75DD86 pKa = 4.46VEE88 pKa = 4.82FKK90 pKa = 10.42NCAIQVSFSDD100 pKa = 4.27PEE102 pKa = 3.93ITLALYY108 pKa = 10.52DD109 pKa = 4.38HH110 pKa = 7.12LALYY114 pKa = 7.08QQCVIQFTSFAEE126 pKa = 4.52GTTTCEE132 pKa = 3.61SWGPTPIPLDD142 pKa = 3.52LCLCDD147 pKa = 4.13EE148 pKa = 5.03GGLNKK153 pKa = 10.33CAVSGAKK160 pKa = 9.1TIHH163 pKa = 5.57VFYY166 pKa = 10.11PSGPAVSAAKK176 pKa = 9.63FQKK179 pKa = 10.49NQTLLITTDD188 pKa = 3.17DD189 pKa = 3.87CGRR192 pKa = 11.84TLSTASTASPISDD205 pKa = 3.81SEE207 pKa = 4.2KK208 pKa = 10.3HH209 pKa = 6.57RR210 pKa = 11.84SICFQLSPKK219 pKa = 9.71PEE221 pKa = 4.5NIGQHH226 pKa = 4.82ILRR229 pKa = 11.84VIRR232 pKa = 11.84QPLTAISRR240 pKa = 11.84QGPFSYY246 pKa = 11.14LNDD249 pKa = 3.34THH251 pKa = 6.82TLLLPGPVSQEE262 pKa = 3.52QHH264 pKa = 5.6DD265 pKa = 4.36TYY267 pKa = 11.26DD268 pKa = 3.42GSGEE272 pKa = 4.04MEE274 pKa = 4.6PNWFNYY280 pKa = 8.29TYY282 pKa = 10.94QDD284 pKa = 4.06DD285 pKa = 4.29LTLALYY291 pKa = 10.68SGDD294 pKa = 3.99LLDD297 pKa = 4.8VLGDD301 pKa = 3.52DD302 pKa = 3.54TVFFVFPDD310 pKa = 4.4SICVNYY316 pKa = 9.68TSHH319 pKa = 7.48PYY321 pKa = 10.3SEE323 pKa = 4.72TSAEE327 pKa = 4.13YY328 pKa = 10.73GDD330 pKa = 3.82SSEE333 pKa = 4.05ALEE336 pKa = 4.1YY337 pKa = 10.7TIGRR341 pKa = 11.84VDD343 pKa = 3.54ACTLTAIQTQINGTVIYY360 pKa = 9.81DD361 pKa = 3.62VPCEE365 pKa = 3.9EE366 pKa = 4.55GQTCFGYY373 pKa = 10.87VYY375 pKa = 10.6GDD377 pKa = 3.4RR378 pKa = 11.84MKK380 pKa = 10.59LTVAHH385 pKa = 6.93PGYY388 pKa = 6.98TTQEE392 pKa = 4.09HH393 pKa = 6.71NYY395 pKa = 9.31GNYY398 pKa = 9.6VEE400 pKa = 5.55LDD402 pKa = 3.89AYY404 pKa = 11.22DD405 pKa = 4.3MITPEE410 pKa = 4.52PGSPSKK416 pKa = 10.81HH417 pKa = 6.29IIWKK421 pKa = 5.75TTLFSDD427 pKa = 4.96LSDD430 pKa = 4.35DD431 pKa = 4.88FVDD434 pKa = 3.62TRR436 pKa = 11.84LNRR439 pKa = 11.84HH440 pKa = 5.45QLDD443 pKa = 4.23GVCDD447 pKa = 4.11DD448 pKa = 4.77EE449 pKa = 4.85GCPGEE454 pKa = 4.99GSWEE458 pKa = 4.24DD459 pKa = 3.33IYY461 pKa = 10.75PTVHH465 pKa = 6.33EE466 pKa = 4.45
Molecular weight: 51.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1R159|A0A0K1R159_9ALPH ICP4-like protein OS=Testudinid alphaherpesvirus 3 OX=2560801 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.66EE2 pKa = 5.31SGGNGRR8 pKa = 11.84GRR10 pKa = 11.84PRR12 pKa = 11.84GRR14 pKa = 11.84PTRR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84GRR22 pKa = 11.84RR23 pKa = 11.84GPSPPRR29 pKa = 11.84TRR31 pKa = 11.84SRR33 pKa = 11.84SPLRR37 pKa = 11.84HH38 pKa = 6.02GPDD41 pKa = 3.26SPEE44 pKa = 4.37PGPSGSGMVPRR55 pKa = 11.84LVRR58 pKa = 11.84PSPGDD63 pKa = 3.36PEE65 pKa = 4.16QAHH68 pKa = 6.86PGSPARR74 pKa = 11.84QQPRR78 pKa = 11.84DD79 pKa = 3.61NGGSSDD85 pKa = 3.8STVSLSNPEE94 pKa = 4.01SDD96 pKa = 3.33SGQITSSDD104 pKa = 3.75SGDD107 pKa = 3.69EE108 pKa = 3.9AHH110 pKa = 7.37SPGLVQGPVSPGPTPFPRR128 pKa = 11.84PGTLPVEE135 pKa = 4.49VGDD138 pKa = 4.07VSSSTSVQAPPTGNQGRR155 pKa = 11.84SPGEE159 pKa = 3.86VAPDD163 pKa = 3.79PPHH166 pKa = 6.11IPRR169 pKa = 11.84WVRR172 pKa = 11.84LAGQPNIGPMPPIARR187 pKa = 11.84DD188 pKa = 3.38QRR190 pKa = 11.84ISEE193 pKa = 4.34DD194 pKa = 4.26HH195 pKa = 6.92PPADD199 pKa = 3.77PGNYY203 pKa = 9.35VFIIYY208 pKa = 9.17TGG210 pKa = 3.16
MM1 pKa = 7.66EE2 pKa = 5.31SGGNGRR8 pKa = 11.84GRR10 pKa = 11.84PRR12 pKa = 11.84GRR14 pKa = 11.84PTRR17 pKa = 11.84RR18 pKa = 11.84SRR20 pKa = 11.84GRR22 pKa = 11.84RR23 pKa = 11.84GPSPPRR29 pKa = 11.84TRR31 pKa = 11.84SRR33 pKa = 11.84SPLRR37 pKa = 11.84HH38 pKa = 6.02GPDD41 pKa = 3.26SPEE44 pKa = 4.37PGPSGSGMVPRR55 pKa = 11.84LVRR58 pKa = 11.84PSPGDD63 pKa = 3.36PEE65 pKa = 4.16QAHH68 pKa = 6.86PGSPARR74 pKa = 11.84QQPRR78 pKa = 11.84DD79 pKa = 3.61NGGSSDD85 pKa = 3.8STVSLSNPEE94 pKa = 4.01SDD96 pKa = 3.33SGQITSSDD104 pKa = 3.75SGDD107 pKa = 3.69EE108 pKa = 3.9AHH110 pKa = 7.37SPGLVQGPVSPGPTPFPRR128 pKa = 11.84PGTLPVEE135 pKa = 4.49VGDD138 pKa = 4.07VSSSTSVQAPPTGNQGRR155 pKa = 11.84SPGEE159 pKa = 3.86VAPDD163 pKa = 3.79PPHH166 pKa = 6.11IPRR169 pKa = 11.84WVRR172 pKa = 11.84LAGQPNIGPMPPIARR187 pKa = 11.84DD188 pKa = 3.38QRR190 pKa = 11.84ISEE193 pKa = 4.34DD194 pKa = 4.26HH195 pKa = 6.92PPADD199 pKa = 3.77PGNYY203 pKa = 9.35VFIIYY208 pKa = 9.17TGG210 pKa = 3.16
Molecular weight: 21.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
42401 |
79 |
2574 |
424.0 |
48.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.021 ± 0.201 | 2.323 ± 0.166 |
5.46 ± 0.14 | 5.604 ± 0.154 |
4.328 ± 0.151 | 5.243 ± 0.176 |
2.535 ± 0.092 | 6.141 ± 0.17 |
4.627 ± 0.173 | 9.698 ± 0.185 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.571 ± 0.097 | 4.795 ± 0.136 |
5.354 ± 0.225 | 3.266 ± 0.116 |
5.887 ± 0.171 | 7.493 ± 0.176 |
6.762 ± 0.143 | 6.415 ± 0.129 |
1.149 ± 0.087 | 4.33 ± 0.148 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
