
Enteractinococcus helveticum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Enteractinococcus
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
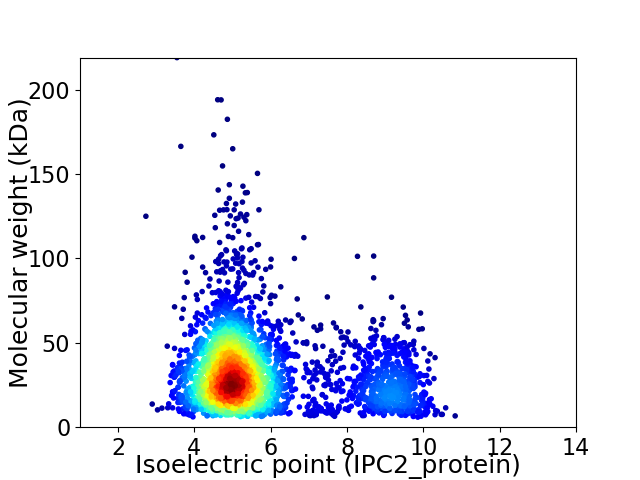
Virtual 2D-PAGE plot for 3201 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
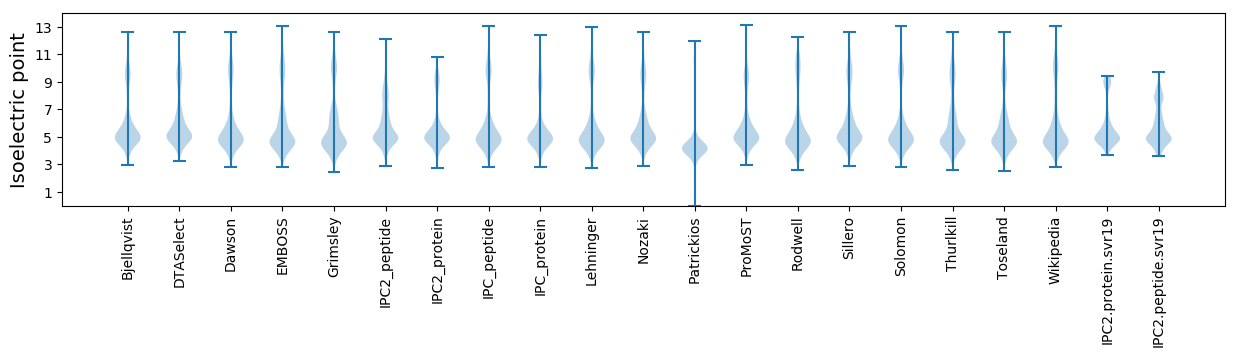
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B7LXW3|A0A1B7LXW3_9MICC Uncharacterized protein OS=Enteractinococcus helveticum OX=1837282 GN=A6F49_12085 PE=4 SV=1
MM1 pKa = 7.62SKK3 pKa = 10.27PIKK6 pKa = 10.16QYY8 pKa = 10.02WPIAATLGLIAVIVLALLRR27 pKa = 11.84PWSTPKK33 pKa = 10.29TEE35 pKa = 4.41LTFEE39 pKa = 4.66LSPTQVDD46 pKa = 3.45IAIDD50 pKa = 3.73SQDD53 pKa = 3.7YY54 pKa = 10.78GSVASGEE61 pKa = 4.09MLEE64 pKa = 4.51VEE66 pKa = 4.48LSGDD70 pKa = 3.93VIVEE74 pKa = 3.98ASRR77 pKa = 11.84EE78 pKa = 4.17GFTPEE83 pKa = 4.25SLPLTVDD90 pKa = 2.99PGEE93 pKa = 4.21AQTVYY98 pKa = 10.67FGLVPEE104 pKa = 4.41TPEE107 pKa = 4.22AEE109 pKa = 4.18EE110 pKa = 5.26LVDD113 pKa = 3.89EE114 pKa = 4.32QDD116 pKa = 3.66SVVDD120 pKa = 3.71EE121 pKa = 4.06QVITEE126 pKa = 4.38EE127 pKa = 3.92YY128 pKa = 9.59HH129 pKa = 7.4DD130 pKa = 3.98RR131 pKa = 11.84AEE133 pKa = 4.47GMFDD137 pKa = 3.12KK138 pKa = 11.05HH139 pKa = 6.74PILSDD144 pKa = 3.86LPQEE148 pKa = 4.19DD149 pKa = 4.21QYY151 pKa = 11.97YY152 pKa = 10.49GAYY155 pKa = 9.69QGIPEE160 pKa = 4.5GDD162 pKa = 3.07AHH164 pKa = 7.14DD165 pKa = 4.24FAIHH169 pKa = 5.41LHH171 pKa = 6.45LYY173 pKa = 9.31VGHH176 pKa = 6.51EE177 pKa = 3.92AQGRR181 pKa = 11.84EE182 pKa = 4.13DD183 pKa = 3.91FEE185 pKa = 4.62VWMDD189 pKa = 3.65DD190 pKa = 3.69AGYY193 pKa = 8.23STSNYY198 pKa = 10.16QIIEE202 pKa = 4.54DD203 pKa = 4.5IKK205 pKa = 11.52DD206 pKa = 3.45EE207 pKa = 4.77DD208 pKa = 4.31PPVIPDD214 pKa = 4.54DD215 pKa = 4.53GPSYY219 pKa = 9.9TALQDD224 pKa = 3.7MNPGDD229 pKa = 3.81ISMPTADD236 pKa = 3.99NKK238 pKa = 11.26GLGVDD243 pKa = 3.97ALALHH248 pKa = 6.48FAEE251 pKa = 6.35VSTTWDD257 pKa = 3.1TAEE260 pKa = 4.92DD261 pKa = 3.63VHH263 pKa = 6.47HH264 pKa = 7.01TDD266 pKa = 2.84GLIRR270 pKa = 11.84AKK272 pKa = 10.55PLMTHH277 pKa = 7.21DD278 pKa = 4.42LADD281 pKa = 4.49SIEE284 pKa = 4.34MPHH287 pKa = 6.87RR288 pKa = 11.84PTPSQSWWTAYY299 pKa = 7.59EE300 pKa = 4.29HH301 pKa = 6.17EE302 pKa = 4.66AKK304 pKa = 10.58SYY306 pKa = 10.5VWLHH310 pKa = 6.97DD311 pKa = 3.65YY312 pKa = 11.01SSQHH316 pKa = 5.59YY317 pKa = 9.23RR318 pKa = 11.84DD319 pKa = 3.54NSTVEE324 pKa = 4.15MQVCWAWVADD334 pKa = 3.77EE335 pKa = 4.78MEE337 pKa = 4.51PVVDD341 pKa = 5.05GPRR344 pKa = 11.84TLEE347 pKa = 3.94ATVDD351 pKa = 3.58TTGSDD356 pKa = 3.61NIISDD361 pKa = 4.26FFYY364 pKa = 10.73EE365 pKa = 4.87DD366 pKa = 3.79PDD368 pKa = 4.43DD369 pKa = 5.07FVDD372 pKa = 5.29SSASPCIPEE381 pKa = 4.48DD382 pKa = 3.46ASS384 pKa = 3.41
MM1 pKa = 7.62SKK3 pKa = 10.27PIKK6 pKa = 10.16QYY8 pKa = 10.02WPIAATLGLIAVIVLALLRR27 pKa = 11.84PWSTPKK33 pKa = 10.29TEE35 pKa = 4.41LTFEE39 pKa = 4.66LSPTQVDD46 pKa = 3.45IAIDD50 pKa = 3.73SQDD53 pKa = 3.7YY54 pKa = 10.78GSVASGEE61 pKa = 4.09MLEE64 pKa = 4.51VEE66 pKa = 4.48LSGDD70 pKa = 3.93VIVEE74 pKa = 3.98ASRR77 pKa = 11.84EE78 pKa = 4.17GFTPEE83 pKa = 4.25SLPLTVDD90 pKa = 2.99PGEE93 pKa = 4.21AQTVYY98 pKa = 10.67FGLVPEE104 pKa = 4.41TPEE107 pKa = 4.22AEE109 pKa = 4.18EE110 pKa = 5.26LVDD113 pKa = 3.89EE114 pKa = 4.32QDD116 pKa = 3.66SVVDD120 pKa = 3.71EE121 pKa = 4.06QVITEE126 pKa = 4.38EE127 pKa = 3.92YY128 pKa = 9.59HH129 pKa = 7.4DD130 pKa = 3.98RR131 pKa = 11.84AEE133 pKa = 4.47GMFDD137 pKa = 3.12KK138 pKa = 11.05HH139 pKa = 6.74PILSDD144 pKa = 3.86LPQEE148 pKa = 4.19DD149 pKa = 4.21QYY151 pKa = 11.97YY152 pKa = 10.49GAYY155 pKa = 9.69QGIPEE160 pKa = 4.5GDD162 pKa = 3.07AHH164 pKa = 7.14DD165 pKa = 4.24FAIHH169 pKa = 5.41LHH171 pKa = 6.45LYY173 pKa = 9.31VGHH176 pKa = 6.51EE177 pKa = 3.92AQGRR181 pKa = 11.84EE182 pKa = 4.13DD183 pKa = 3.91FEE185 pKa = 4.62VWMDD189 pKa = 3.65DD190 pKa = 3.69AGYY193 pKa = 8.23STSNYY198 pKa = 10.16QIIEE202 pKa = 4.54DD203 pKa = 4.5IKK205 pKa = 11.52DD206 pKa = 3.45EE207 pKa = 4.77DD208 pKa = 4.31PPVIPDD214 pKa = 4.54DD215 pKa = 4.53GPSYY219 pKa = 9.9TALQDD224 pKa = 3.7MNPGDD229 pKa = 3.81ISMPTADD236 pKa = 3.99NKK238 pKa = 11.26GLGVDD243 pKa = 3.97ALALHH248 pKa = 6.48FAEE251 pKa = 6.35VSTTWDD257 pKa = 3.1TAEE260 pKa = 4.92DD261 pKa = 3.63VHH263 pKa = 6.47HH264 pKa = 7.01TDD266 pKa = 2.84GLIRR270 pKa = 11.84AKK272 pKa = 10.55PLMTHH277 pKa = 7.21DD278 pKa = 4.42LADD281 pKa = 4.49SIEE284 pKa = 4.34MPHH287 pKa = 6.87RR288 pKa = 11.84PTPSQSWWTAYY299 pKa = 7.59EE300 pKa = 4.29HH301 pKa = 6.17EE302 pKa = 4.66AKK304 pKa = 10.58SYY306 pKa = 10.5VWLHH310 pKa = 6.97DD311 pKa = 3.65YY312 pKa = 11.01SSQHH316 pKa = 5.59YY317 pKa = 9.23RR318 pKa = 11.84DD319 pKa = 3.54NSTVEE324 pKa = 4.15MQVCWAWVADD334 pKa = 3.77EE335 pKa = 4.78MEE337 pKa = 4.51PVVDD341 pKa = 5.05GPRR344 pKa = 11.84TLEE347 pKa = 3.94ATVDD351 pKa = 3.58TTGSDD356 pKa = 3.61NIISDD361 pKa = 4.26FFYY364 pKa = 10.73EE365 pKa = 4.87DD366 pKa = 3.79PDD368 pKa = 4.43DD369 pKa = 5.07FVDD372 pKa = 5.29SSASPCIPEE381 pKa = 4.48DD382 pKa = 3.46ASS384 pKa = 3.41
Molecular weight: 42.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B7LWN3|A0A1B7LWN3_9MICC Amino_oxidase domain-containing protein OS=Enteractinococcus helveticum OX=1837282 GN=A6F49_16625 PE=4 SV=1
MM1 pKa = 7.72ASNHH5 pKa = 5.14NRR7 pKa = 11.84SVGAFFAGVFMVNSLGHH24 pKa = 6.53FATAIAGKK32 pKa = 8.45QHH34 pKa = 6.47LTPLAGRR41 pKa = 11.84RR42 pKa = 11.84SGPAVNALWGAMNLAGGLALARR64 pKa = 11.84RR65 pKa = 4.28
MM1 pKa = 7.72ASNHH5 pKa = 5.14NRR7 pKa = 11.84SVGAFFAGVFMVNSLGHH24 pKa = 6.53FATAIAGKK32 pKa = 8.45QHH34 pKa = 6.47LTPLAGRR41 pKa = 11.84RR42 pKa = 11.84SGPAVNALWGAMNLAGGLALARR64 pKa = 11.84RR65 pKa = 4.28
Molecular weight: 6.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1011829 |
55 |
2016 |
316.1 |
34.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.315 ± 0.045 | 0.508 ± 0.009 |
6.169 ± 0.048 | 6.131 ± 0.048 |
3.319 ± 0.025 | 7.854 ± 0.037 |
2.445 ± 0.024 | 5.453 ± 0.034 |
2.747 ± 0.032 | 9.713 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.36 ± 0.02 | 2.859 ± 0.025 |
4.978 ± 0.032 | 4.095 ± 0.03 |
5.899 ± 0.042 | 5.706 ± 0.032 |
6.631 ± 0.034 | 7.973 ± 0.033 |
1.471 ± 0.019 | 2.373 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
