
Bifidobacterium choloepi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Bifidobacteriales; Bifidobacteriaceae; Bifidobacterium
Average proteome isoelectric point is 5.75
Get precalculated fractions of proteins
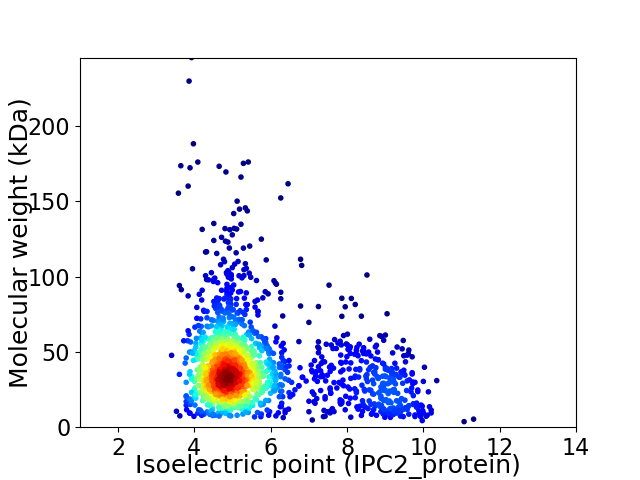
Virtual 2D-PAGE plot for 1599 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
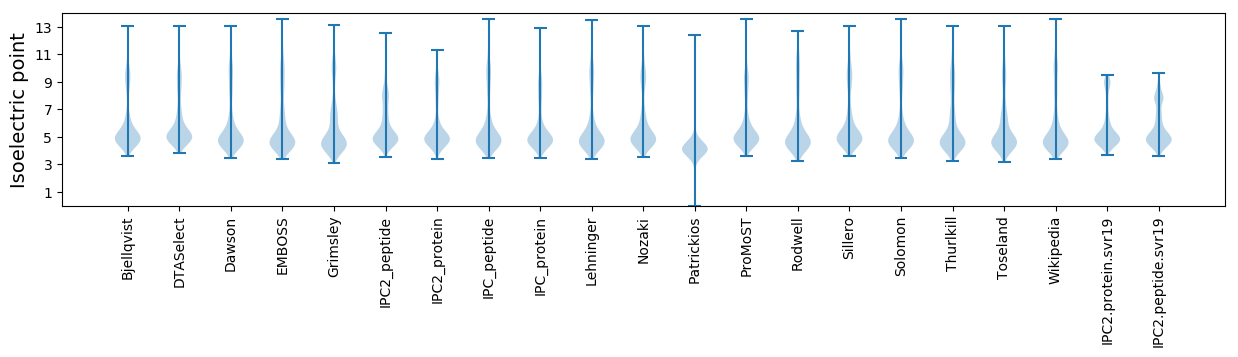
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I5MYS2|A0A6I5MYS2_9BIFI Esterase family protein OS=Bifidobacterium choloepi OX=2614131 GN=F6S87_03990 PE=4 SV=1
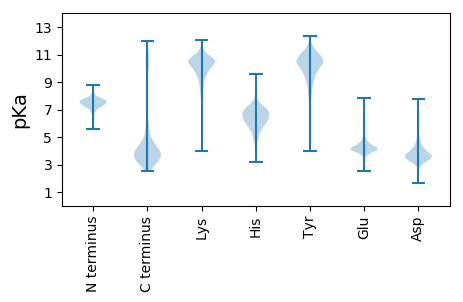
MM1 pKa = 7.59GRR3 pKa = 11.84LTKK6 pKa = 10.74LCTAVAASLALAGVAVGPAFAGEE29 pKa = 4.22SSGGSSTTTSTIDD42 pKa = 3.39AGNGTITVDD51 pKa = 2.83YY52 pKa = 9.48HH53 pKa = 4.29YY54 pKa = 9.24TYY56 pKa = 10.24TSSAGEE62 pKa = 3.8QTQFPVKK69 pKa = 9.75GANVMLYY76 pKa = 10.22KK77 pKa = 10.63AAGWNDD83 pKa = 3.28SGDD86 pKa = 3.7LVLTGDD92 pKa = 4.81FAALCEE98 pKa = 4.3GASLDD103 pKa = 4.17CSAITDD109 pKa = 3.73TGRR112 pKa = 11.84LADD115 pKa = 4.4NDD117 pKa = 4.17TQRR120 pKa = 11.84DD121 pKa = 3.86LATSLLTALATSDD134 pKa = 3.98IAFDD138 pKa = 3.69TQDD141 pKa = 3.21AVIVSDD147 pKa = 3.69GTARR151 pKa = 11.84YY152 pKa = 9.79VDD154 pKa = 3.95ADD156 pKa = 3.88GEE158 pKa = 4.4ATDD161 pKa = 5.53DD162 pKa = 3.95VAFTGLGDD170 pKa = 3.63ALYY173 pKa = 9.76FIRR176 pKa = 11.84VGGQILDD183 pKa = 3.58ADD185 pKa = 4.44AYY187 pKa = 10.12SHH189 pKa = 7.0LMYY192 pKa = 10.84CNEE195 pKa = 3.68TAMFIDD201 pKa = 4.71LKK203 pKa = 10.94SAVSGDD209 pKa = 3.82DD210 pKa = 3.26VAGTVSRR217 pKa = 11.84TVSLEE222 pKa = 3.78PKK224 pKa = 10.07ASCTLGPDD232 pKa = 4.5PIHH235 pKa = 5.7VTKK238 pKa = 10.72VWDD241 pKa = 4.97DD242 pKa = 3.59GDD244 pKa = 3.9STARR248 pKa = 11.84PSSVDD253 pKa = 2.91VTLYY257 pKa = 10.94GRR259 pKa = 11.84YY260 pKa = 7.97TDD262 pKa = 3.56IEE264 pKa = 4.3GNEE267 pKa = 3.99VVEE270 pKa = 4.75TIEE273 pKa = 4.21TQTLSADD280 pKa = 3.75NDD282 pKa = 3.45WTYY285 pKa = 11.35VWSGLPDD292 pKa = 3.92DD293 pKa = 4.28YY294 pKa = 11.79YY295 pKa = 11.01EE296 pKa = 4.45YY297 pKa = 10.63FVQEE301 pKa = 3.62VDD303 pKa = 3.63VPDD306 pKa = 5.31GYY308 pKa = 9.35TVSYY312 pKa = 10.81SRR314 pKa = 11.84TDD316 pKa = 3.22VTEE319 pKa = 5.13EE320 pKa = 3.94SATWTVTNTSEE331 pKa = 4.89SEE333 pKa = 4.03SSTPGDD339 pKa = 3.51SEE341 pKa = 4.77TPGTSDD347 pKa = 4.25SDD349 pKa = 4.26DD350 pKa = 3.35GTTPGTSGSTGSTSASASATATSTTPAQTGANIALVALVAVSALMLGVLLIVEE403 pKa = 4.69IRR405 pKa = 11.84RR406 pKa = 11.84ASKK409 pKa = 10.42KK410 pKa = 10.41AA411 pKa = 3.17
MM1 pKa = 7.59GRR3 pKa = 11.84LTKK6 pKa = 10.74LCTAVAASLALAGVAVGPAFAGEE29 pKa = 4.22SSGGSSTTTSTIDD42 pKa = 3.39AGNGTITVDD51 pKa = 2.83YY52 pKa = 9.48HH53 pKa = 4.29YY54 pKa = 9.24TYY56 pKa = 10.24TSSAGEE62 pKa = 3.8QTQFPVKK69 pKa = 9.75GANVMLYY76 pKa = 10.22KK77 pKa = 10.63AAGWNDD83 pKa = 3.28SGDD86 pKa = 3.7LVLTGDD92 pKa = 4.81FAALCEE98 pKa = 4.3GASLDD103 pKa = 4.17CSAITDD109 pKa = 3.73TGRR112 pKa = 11.84LADD115 pKa = 4.4NDD117 pKa = 4.17TQRR120 pKa = 11.84DD121 pKa = 3.86LATSLLTALATSDD134 pKa = 3.98IAFDD138 pKa = 3.69TQDD141 pKa = 3.21AVIVSDD147 pKa = 3.69GTARR151 pKa = 11.84YY152 pKa = 9.79VDD154 pKa = 3.95ADD156 pKa = 3.88GEE158 pKa = 4.4ATDD161 pKa = 5.53DD162 pKa = 3.95VAFTGLGDD170 pKa = 3.63ALYY173 pKa = 9.76FIRR176 pKa = 11.84VGGQILDD183 pKa = 3.58ADD185 pKa = 4.44AYY187 pKa = 10.12SHH189 pKa = 7.0LMYY192 pKa = 10.84CNEE195 pKa = 3.68TAMFIDD201 pKa = 4.71LKK203 pKa = 10.94SAVSGDD209 pKa = 3.82DD210 pKa = 3.26VAGTVSRR217 pKa = 11.84TVSLEE222 pKa = 3.78PKK224 pKa = 10.07ASCTLGPDD232 pKa = 4.5PIHH235 pKa = 5.7VTKK238 pKa = 10.72VWDD241 pKa = 4.97DD242 pKa = 3.59GDD244 pKa = 3.9STARR248 pKa = 11.84PSSVDD253 pKa = 2.91VTLYY257 pKa = 10.94GRR259 pKa = 11.84YY260 pKa = 7.97TDD262 pKa = 3.56IEE264 pKa = 4.3GNEE267 pKa = 3.99VVEE270 pKa = 4.75TIEE273 pKa = 4.21TQTLSADD280 pKa = 3.75NDD282 pKa = 3.45WTYY285 pKa = 11.35VWSGLPDD292 pKa = 3.92DD293 pKa = 4.28YY294 pKa = 11.79YY295 pKa = 11.01EE296 pKa = 4.45YY297 pKa = 10.63FVQEE301 pKa = 3.62VDD303 pKa = 3.63VPDD306 pKa = 5.31GYY308 pKa = 9.35TVSYY312 pKa = 10.81SRR314 pKa = 11.84TDD316 pKa = 3.22VTEE319 pKa = 5.13EE320 pKa = 3.94SATWTVTNTSEE331 pKa = 4.89SEE333 pKa = 4.03SSTPGDD339 pKa = 3.51SEE341 pKa = 4.77TPGTSDD347 pKa = 4.25SDD349 pKa = 4.26DD350 pKa = 3.35GTTPGTSGSTGSTSASASATATSTTPAQTGANIALVALVAVSALMLGVLLIVEE403 pKa = 4.69IRR405 pKa = 11.84RR406 pKa = 11.84ASKK409 pKa = 10.42KK410 pKa = 10.41AA411 pKa = 3.17
Molecular weight: 42.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I5NMU5|A0A6I5NMU5_9BIFI DNA helicase OS=Bifidobacterium choloepi OX=2614131 GN=F6S87_05435 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 4.79MKK15 pKa = 9.36HH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84SGRR28 pKa = 11.84ALINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84KK40 pKa = 8.87SLSAA44 pKa = 3.86
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84HH13 pKa = 4.79MKK15 pKa = 9.36HH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TRR25 pKa = 11.84SGRR28 pKa = 11.84ALINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.08GRR39 pKa = 11.84KK40 pKa = 8.87SLSAA44 pKa = 3.86
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
615902 |
29 |
2363 |
385.2 |
41.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.338 ± 0.078 | 0.912 ± 0.016 |
7.181 ± 0.052 | 5.793 ± 0.051 |
3.483 ± 0.036 | 8.256 ± 0.05 |
2.055 ± 0.03 | 5.008 ± 0.046 |
3.526 ± 0.055 | 8.717 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.571 ± 0.026 | 3.19 ± 0.04 |
4.609 ± 0.044 | 3.055 ± 0.031 |
5.968 ± 0.07 | 5.97 ± 0.063 |
6.244 ± 0.071 | 8.075 ± 0.055 |
1.335 ± 0.026 | 2.715 ± 0.039 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
