
fungal sp. No.14919
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; unclassified Fungi
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
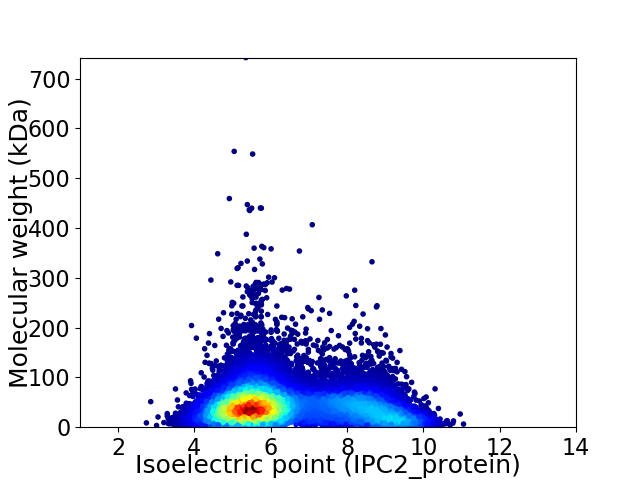
Virtual 2D-PAGE plot for 14332 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1V1TMA7|A0A1V1TMA7_9FUNG Uncharacterized protein OS=fungal sp. No.14919 OX=1813822 GN=ANO14919_114450 PE=4 SV=1
MM1 pKa = 7.22SLSMSPLWLSVDD13 pKa = 3.51YY14 pKa = 11.29CDD16 pKa = 4.64INVTITTQADD26 pKa = 3.6IEE28 pKa = 4.58SLHH31 pKa = 5.12ATPYY35 pKa = 9.66EE36 pKa = 4.06FCFNNVTVANAEE48 pKa = 4.08GTLTFDD54 pKa = 5.06NFTQVNGLYY63 pKa = 10.45VQDD66 pKa = 4.23SPNLEE71 pKa = 3.89ALSFPDD77 pKa = 4.62LGRR80 pKa = 11.84LSALGVYY87 pKa = 9.81DD88 pKa = 5.63ALALSNISLPEE99 pKa = 3.87IQSVGLLPFGGGGWSVPPPWPGAEE123 pKa = 3.92DD124 pKa = 3.49RR125 pKa = 11.84SLIGVTINNAPALKK139 pKa = 9.79TIDD142 pKa = 3.9FDD144 pKa = 4.11FLSGFFTLEE153 pKa = 3.72LVGADD158 pKa = 3.49GLTDD162 pKa = 4.38FGPTGYY168 pKa = 10.62DD169 pKa = 2.86EE170 pKa = 5.99GILRR174 pKa = 11.84RR175 pKa = 11.84INSSDD180 pKa = 3.27TLSVDD185 pKa = 3.36GCFNLANVQFARR197 pKa = 11.84FVQITGRR204 pKa = 11.84TDD206 pKa = 3.17CEE208 pKa = 3.9YY209 pKa = 11.73LMLNYY214 pKa = 9.99HH215 pKa = 5.83SAYY218 pKa = 10.67NLTLIDD224 pKa = 3.85TAHH227 pKa = 6.3SRR229 pKa = 11.84LDD231 pKa = 3.17ILAPFAVNGTLVADD245 pKa = 4.35SLQLSPEE252 pKa = 4.09NSSSYY257 pKa = 11.01DD258 pKa = 3.44PQPLYY263 pKa = 10.69FISSIGEE270 pKa = 4.03DD271 pKa = 3.04AHH273 pKa = 7.01LLSGANVDD281 pKa = 3.91LSLDD285 pKa = 3.64EE286 pKa = 4.78VEE288 pKa = 4.55TLGGSLVASNNKK300 pKa = 9.25NCTFSFDD307 pKa = 3.56KK308 pKa = 10.96LSEE311 pKa = 4.25VKK313 pKa = 10.86GNISMTDD320 pKa = 3.42NPDD323 pKa = 3.23SALPWFPDD331 pKa = 3.14LRR333 pKa = 11.84RR334 pKa = 11.84AANIEE339 pKa = 3.62LRR341 pKa = 11.84GNINTSHH348 pKa = 6.64GPNIFPALTTVPGTVTIEE366 pKa = 3.62AWNSDD371 pKa = 4.19FNCSQLVKK379 pKa = 10.6QMQAGIIQNLVCNGTDD395 pKa = 3.3GTQGDD400 pKa = 4.32TSGSPSSPAGGAWTVLGIAAGIMVLQNTIPVWW432 pKa = 3.47
MM1 pKa = 7.22SLSMSPLWLSVDD13 pKa = 3.51YY14 pKa = 11.29CDD16 pKa = 4.64INVTITTQADD26 pKa = 3.6IEE28 pKa = 4.58SLHH31 pKa = 5.12ATPYY35 pKa = 9.66EE36 pKa = 4.06FCFNNVTVANAEE48 pKa = 4.08GTLTFDD54 pKa = 5.06NFTQVNGLYY63 pKa = 10.45VQDD66 pKa = 4.23SPNLEE71 pKa = 3.89ALSFPDD77 pKa = 4.62LGRR80 pKa = 11.84LSALGVYY87 pKa = 9.81DD88 pKa = 5.63ALALSNISLPEE99 pKa = 3.87IQSVGLLPFGGGGWSVPPPWPGAEE123 pKa = 3.92DD124 pKa = 3.49RR125 pKa = 11.84SLIGVTINNAPALKK139 pKa = 9.79TIDD142 pKa = 3.9FDD144 pKa = 4.11FLSGFFTLEE153 pKa = 3.72LVGADD158 pKa = 3.49GLTDD162 pKa = 4.38FGPTGYY168 pKa = 10.62DD169 pKa = 2.86EE170 pKa = 5.99GILRR174 pKa = 11.84RR175 pKa = 11.84INSSDD180 pKa = 3.27TLSVDD185 pKa = 3.36GCFNLANVQFARR197 pKa = 11.84FVQITGRR204 pKa = 11.84TDD206 pKa = 3.17CEE208 pKa = 3.9YY209 pKa = 11.73LMLNYY214 pKa = 9.99HH215 pKa = 5.83SAYY218 pKa = 10.67NLTLIDD224 pKa = 3.85TAHH227 pKa = 6.3SRR229 pKa = 11.84LDD231 pKa = 3.17ILAPFAVNGTLVADD245 pKa = 4.35SLQLSPEE252 pKa = 4.09NSSSYY257 pKa = 11.01DD258 pKa = 3.44PQPLYY263 pKa = 10.69FISSIGEE270 pKa = 4.03DD271 pKa = 3.04AHH273 pKa = 7.01LLSGANVDD281 pKa = 3.91LSLDD285 pKa = 3.64EE286 pKa = 4.78VEE288 pKa = 4.55TLGGSLVASNNKK300 pKa = 9.25NCTFSFDD307 pKa = 3.56KK308 pKa = 10.96LSEE311 pKa = 4.25VKK313 pKa = 10.86GNISMTDD320 pKa = 3.42NPDD323 pKa = 3.23SALPWFPDD331 pKa = 3.14LRR333 pKa = 11.84RR334 pKa = 11.84AANIEE339 pKa = 3.62LRR341 pKa = 11.84GNINTSHH348 pKa = 6.64GPNIFPALTTVPGTVTIEE366 pKa = 3.62AWNSDD371 pKa = 4.19FNCSQLVKK379 pKa = 10.6QMQAGIIQNLVCNGTDD395 pKa = 3.3GTQGDD400 pKa = 4.32TSGSPSSPAGGAWTVLGIAAGIMVLQNTIPVWW432 pKa = 3.47
Molecular weight: 46.19 kDa
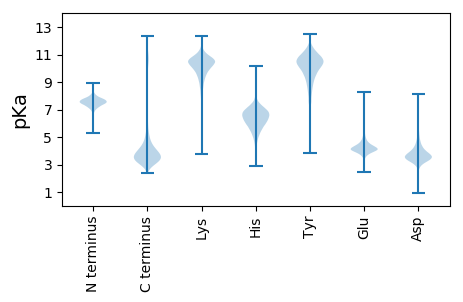
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V1T950|A0A1V1T950_9FUNG Uncharacterized protein OS=fungal sp. No.14919 OX=1813822 GN=ANO14919_068420 PE=4 SV=1
MM1 pKa = 6.57VTGPVPILGLPPGPPLPGLPLGTCASIGITTGVRR35 pKa = 11.84IATSVGITTGVGITTGVGIATSIGITTTSIRR66 pKa = 11.84IAASVGITTSVRR78 pKa = 11.84ITTSVGITTGVGIATSIGITTGVGIATSIGITTTSIRR115 pKa = 11.84IAASVGITTSVRR127 pKa = 11.84ITTSVGITTGVGIATSIGITTGVGIATSIGITTTSIRR164 pKa = 11.84IAASVGITTTSIRR177 pKa = 11.84IAASVGITTSVRR189 pKa = 11.84ITTSVGITTGVGIATSIGITTGVGIATSIGITTTSIRR226 pKa = 11.84IAASVGITTGVRR238 pKa = 11.84TAAGVGIATSVGITAGVGITTSVGIATSIGTATRR272 pKa = 11.84TPTARR277 pKa = 11.84ATVRR281 pKa = 3.27
MM1 pKa = 6.57VTGPVPILGLPPGPPLPGLPLGTCASIGITTGVRR35 pKa = 11.84IATSVGITTGVGITTGVGIATSIGITTTSIRR66 pKa = 11.84IAASVGITTSVRR78 pKa = 11.84ITTSVGITTGVGIATSIGITTGVGIATSIGITTTSIRR115 pKa = 11.84IAASVGITTSVRR127 pKa = 11.84ITTSVGITTGVGIATSIGITTGVGIATSIGITTTSIRR164 pKa = 11.84IAASVGITTTSIRR177 pKa = 11.84IAASVGITTSVRR189 pKa = 11.84ITTSVGITTGVGIATSIGITTGVGIATSIGITTTSIRR226 pKa = 11.84IAASVGITTGVRR238 pKa = 11.84TAAGVGIATSVGITAGVGITTSVGIATSIGTATRR272 pKa = 11.84TPTARR277 pKa = 11.84ATVRR281 pKa = 3.27
Molecular weight: 26.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6368111 |
29 |
6731 |
444.3 |
49.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.646 ± 0.018 | 1.227 ± 0.007 |
5.788 ± 0.015 | 6.059 ± 0.016 |
3.705 ± 0.013 | 6.974 ± 0.017 |
2.401 ± 0.01 | 5.027 ± 0.016 |
4.579 ± 0.018 | 8.953 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.032 ± 0.008 | 3.72 ± 0.011 |
6.034 ± 0.025 | 3.883 ± 0.014 |
6.234 ± 0.019 | 8.188 ± 0.023 |
6.052 ± 0.018 | 6.153 ± 0.015 |
1.511 ± 0.007 | 2.835 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
