
Pichia kudriavzevii (Yeast) (Issatchenkia orientalis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Pichiaceae; Pichia
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
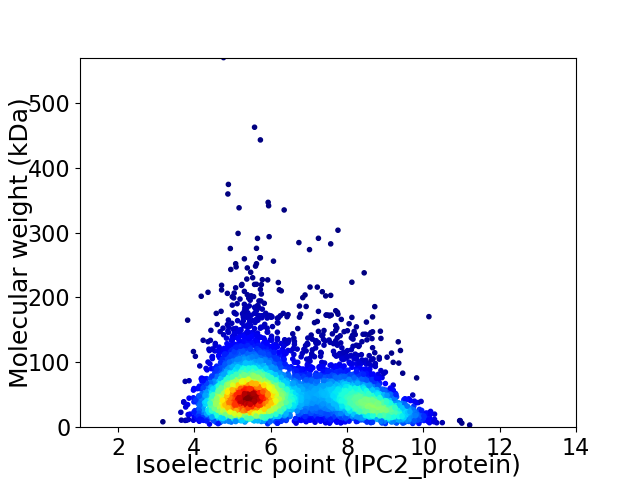
Virtual 2D-PAGE plot for 5146 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U9R0P9|A0A2U9R0P9_PICKU Uncharacterized protein OS=Pichia kudriavzevii OX=4909 GN=C5L36_0B00160 PE=4 SV=1
MM1 pKa = 7.5NFQISLIFFTFLQVILAAPLPYY23 pKa = 10.01YY24 pKa = 9.65VTVTAPDD31 pKa = 3.73TYY33 pKa = 9.5VTTIIPVAEE42 pKa = 4.43VIISDD47 pKa = 3.94GQTTTIQLTTLVTQTSPAADD67 pKa = 3.78PTTLATEE74 pKa = 4.48TNLPADD80 pKa = 3.8PTTEE84 pKa = 4.07NTEE87 pKa = 4.28TTPEE91 pKa = 4.11TTVDD95 pKa = 3.4SVTPATSEE103 pKa = 4.3TTSTPALTTSDD114 pKa = 3.94CTSTSISVDD123 pKa = 3.67SNGVAHH129 pKa = 6.72VWVTLDD135 pKa = 3.15KK136 pKa = 10.26TVLVGTDD143 pKa = 3.59GVPFDD148 pKa = 4.33TQTEE152 pKa = 4.17QLPQATEE159 pKa = 4.44TPVSTSSTQTSVDD172 pKa = 3.39LVGNNNVAAATLSTDD187 pKa = 3.4TTSSFTSATTGSTSPTTQSTPNTTNTSSEE216 pKa = 4.29DD217 pKa = 3.39STSAISTTLATEE229 pKa = 4.41TTSSSSAQDD238 pKa = 3.43SSSEE242 pKa = 4.16TSLSMNTDD250 pKa = 2.93AAGKK254 pKa = 9.56DD255 pKa = 3.53KK256 pKa = 11.2PFVTTWANGEE266 pKa = 4.2VFYY269 pKa = 11.41SSLPFSLVPSEE280 pKa = 4.17EE281 pKa = 4.11VAPNVALTTTTLTNINDD298 pKa = 4.05APTTTALLSLVSSSDD313 pKa = 3.34KK314 pKa = 11.12LLLPTLTVGLPKK326 pKa = 10.62DD327 pKa = 3.68KK328 pKa = 11.13NVNTNMADD336 pKa = 3.51PATVSVTNTKK346 pKa = 10.13QVTTPVIVTTSSTSSTTPTADD367 pKa = 3.44STSSSSPDD375 pKa = 3.76DD376 pKa = 3.65VLQDD380 pKa = 3.79EE381 pKa = 4.63QTSTSAASSSASSDD395 pKa = 3.22SGLLTKK401 pKa = 10.5APYY404 pKa = 10.29SIVYY408 pKa = 9.98SPYY411 pKa = 11.23NNDD414 pKa = 3.59NSCKK418 pKa = 10.39SYY420 pKa = 8.0TTVYY424 pKa = 10.58TDD426 pKa = 4.24LKK428 pKa = 10.7LIASKK433 pKa = 10.58GIKK436 pKa = 9.51EE437 pKa = 3.54IRR439 pKa = 11.84IYY441 pKa = 11.33GNDD444 pKa = 3.6CNYY447 pKa = 8.83LTTVLNVANKK457 pKa = 10.45LGLKK461 pKa = 9.43VNQGFWISQDD471 pKa = 3.19GANSIDD477 pKa = 3.64DD478 pKa = 4.02AVDD481 pKa = 3.21NFITYY486 pKa = 9.85ISSGAAEE493 pKa = 4.22YY494 pKa = 10.7SWDD497 pKa = 3.68LFSYY501 pKa = 8.83ITVGNEE507 pKa = 3.93AIISNYY513 pKa = 9.87CSVDD517 pKa = 3.37DD518 pKa = 5.32LINKK522 pKa = 9.41ISEE525 pKa = 4.37VKK527 pKa = 10.67GKK529 pKa = 10.51LNAAGYY535 pKa = 8.91SGKK538 pKa = 8.4ITTSEE543 pKa = 4.05PPVSFEE549 pKa = 5.56NNPQLCTDD557 pKa = 3.74SEE559 pKa = 4.34IDD561 pKa = 3.56FVGINPHH568 pKa = 6.42SYY570 pKa = 10.21FDD572 pKa = 4.09VYY574 pKa = 11.37SSADD578 pKa = 3.32NSGVFVAGQIEE589 pKa = 4.67IVKK592 pKa = 9.24QYY594 pKa = 10.84CGDD597 pKa = 3.39KK598 pKa = 10.75DD599 pKa = 3.6IVVTEE604 pKa = 4.13TGYY607 pKa = 10.28PSAGNVNGDD616 pKa = 3.31NVPSVEE622 pKa = 4.05NQRR625 pKa = 11.84IAVQSILDD633 pKa = 3.71VVGTDD638 pKa = 3.29VTILSTYY645 pKa = 10.62DD646 pKa = 4.76DD647 pKa = 4.05YY648 pKa = 11.61WKK650 pKa = 10.75QPGPYY655 pKa = 9.74NIEE658 pKa = 3.64QHH660 pKa = 6.21FGIIQLLPSVV670 pKa = 4.02
MM1 pKa = 7.5NFQISLIFFTFLQVILAAPLPYY23 pKa = 10.01YY24 pKa = 9.65VTVTAPDD31 pKa = 3.73TYY33 pKa = 9.5VTTIIPVAEE42 pKa = 4.43VIISDD47 pKa = 3.94GQTTTIQLTTLVTQTSPAADD67 pKa = 3.78PTTLATEE74 pKa = 4.48TNLPADD80 pKa = 3.8PTTEE84 pKa = 4.07NTEE87 pKa = 4.28TTPEE91 pKa = 4.11TTVDD95 pKa = 3.4SVTPATSEE103 pKa = 4.3TTSTPALTTSDD114 pKa = 3.94CTSTSISVDD123 pKa = 3.67SNGVAHH129 pKa = 6.72VWVTLDD135 pKa = 3.15KK136 pKa = 10.26TVLVGTDD143 pKa = 3.59GVPFDD148 pKa = 4.33TQTEE152 pKa = 4.17QLPQATEE159 pKa = 4.44TPVSTSSTQTSVDD172 pKa = 3.39LVGNNNVAAATLSTDD187 pKa = 3.4TTSSFTSATTGSTSPTTQSTPNTTNTSSEE216 pKa = 4.29DD217 pKa = 3.39STSAISTTLATEE229 pKa = 4.41TTSSSSAQDD238 pKa = 3.43SSSEE242 pKa = 4.16TSLSMNTDD250 pKa = 2.93AAGKK254 pKa = 9.56DD255 pKa = 3.53KK256 pKa = 11.2PFVTTWANGEE266 pKa = 4.2VFYY269 pKa = 11.41SSLPFSLVPSEE280 pKa = 4.17EE281 pKa = 4.11VAPNVALTTTTLTNINDD298 pKa = 4.05APTTTALLSLVSSSDD313 pKa = 3.34KK314 pKa = 11.12LLLPTLTVGLPKK326 pKa = 10.62DD327 pKa = 3.68KK328 pKa = 11.13NVNTNMADD336 pKa = 3.51PATVSVTNTKK346 pKa = 10.13QVTTPVIVTTSSTSSTTPTADD367 pKa = 3.44STSSSSPDD375 pKa = 3.76DD376 pKa = 3.65VLQDD380 pKa = 3.79EE381 pKa = 4.63QTSTSAASSSASSDD395 pKa = 3.22SGLLTKK401 pKa = 10.5APYY404 pKa = 10.29SIVYY408 pKa = 9.98SPYY411 pKa = 11.23NNDD414 pKa = 3.59NSCKK418 pKa = 10.39SYY420 pKa = 8.0TTVYY424 pKa = 10.58TDD426 pKa = 4.24LKK428 pKa = 10.7LIASKK433 pKa = 10.58GIKK436 pKa = 9.51EE437 pKa = 3.54IRR439 pKa = 11.84IYY441 pKa = 11.33GNDD444 pKa = 3.6CNYY447 pKa = 8.83LTTVLNVANKK457 pKa = 10.45LGLKK461 pKa = 9.43VNQGFWISQDD471 pKa = 3.19GANSIDD477 pKa = 3.64DD478 pKa = 4.02AVDD481 pKa = 3.21NFITYY486 pKa = 9.85ISSGAAEE493 pKa = 4.22YY494 pKa = 10.7SWDD497 pKa = 3.68LFSYY501 pKa = 8.83ITVGNEE507 pKa = 3.93AIISNYY513 pKa = 9.87CSVDD517 pKa = 3.37DD518 pKa = 5.32LINKK522 pKa = 9.41ISEE525 pKa = 4.37VKK527 pKa = 10.67GKK529 pKa = 10.51LNAAGYY535 pKa = 8.91SGKK538 pKa = 8.4ITTSEE543 pKa = 4.05PPVSFEE549 pKa = 5.56NNPQLCTDD557 pKa = 3.74SEE559 pKa = 4.34IDD561 pKa = 3.56FVGINPHH568 pKa = 6.42SYY570 pKa = 10.21FDD572 pKa = 4.09VYY574 pKa = 11.37SSADD578 pKa = 3.32NSGVFVAGQIEE589 pKa = 4.67IVKK592 pKa = 9.24QYY594 pKa = 10.84CGDD597 pKa = 3.39KK598 pKa = 10.75DD599 pKa = 3.6IVVTEE604 pKa = 4.13TGYY607 pKa = 10.28PSAGNVNGDD616 pKa = 3.31NVPSVEE622 pKa = 4.05NQRR625 pKa = 11.84IAVQSILDD633 pKa = 3.71VVGTDD638 pKa = 3.29VTILSTYY645 pKa = 10.62DD646 pKa = 4.76DD647 pKa = 4.05YY648 pKa = 11.61WKK650 pKa = 10.75QPGPYY655 pKa = 9.74NIEE658 pKa = 3.64QHH660 pKa = 6.21FGIIQLLPSVV670 pKa = 4.02
Molecular weight: 70.62 kDa
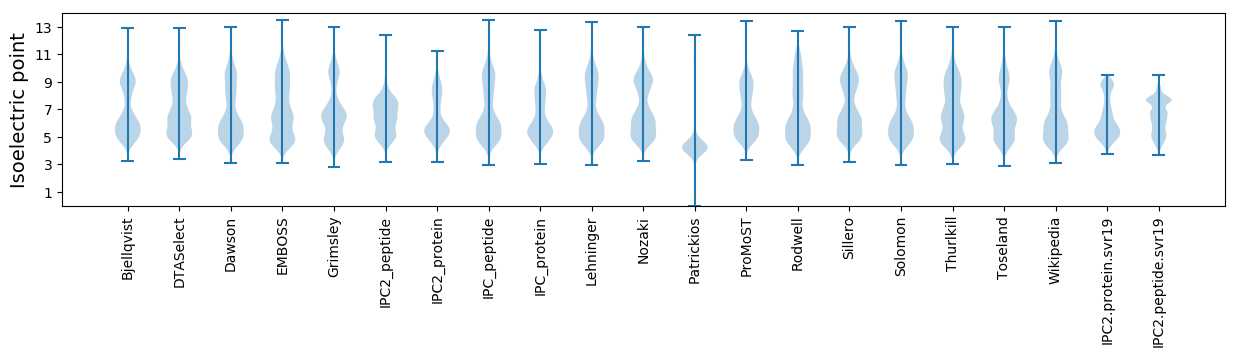
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U9RAN9|A0A2U9RAN9_PICKU TUG-UBL1 domain-containing protein OS=Pichia kudriavzevii OX=4909 GN=C5L36_0E05320 PE=4 SV=1
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.09WRR6 pKa = 11.84KK7 pKa = 8.87KK8 pKa = 8.73RR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.71RR15 pKa = 11.84KK16 pKa = 8.27RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.19VRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 10.98
MM1 pKa = 7.1RR2 pKa = 11.84AKK4 pKa = 9.09WRR6 pKa = 11.84KK7 pKa = 8.87KK8 pKa = 8.73RR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84LKK14 pKa = 9.71RR15 pKa = 11.84KK16 pKa = 8.27RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 9.19VRR21 pKa = 11.84ARR23 pKa = 11.84SKK25 pKa = 10.98
Molecular weight: 3.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2636562 |
25 |
5008 |
512.4 |
57.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.233 ± 0.031 | 1.244 ± 0.012 |
5.993 ± 0.025 | 6.848 ± 0.032 |
4.442 ± 0.024 | 5.195 ± 0.027 |
2.136 ± 0.012 | 6.702 ± 0.022 |
7.348 ± 0.034 | 9.516 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.15 ± 0.012 | 6.111 ± 0.035 |
4.196 ± 0.025 | 3.792 ± 0.028 |
4.34 ± 0.021 | 8.77 ± 0.048 |
5.65 ± 0.028 | 5.83 ± 0.024 |
0.981 ± 0.009 | 3.525 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
