
Tahibacter aquaticus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Rhodanobacteraceae; Tahibacter
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
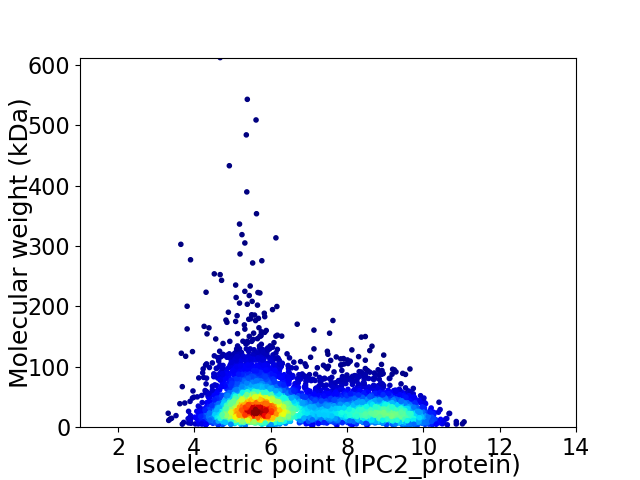
Virtual 2D-PAGE plot for 5664 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R6YSH2|A0A4R6YSH2_9GAMM Diguanylate cyclase (GGDEF)-like protein OS=Tahibacter aquaticus OX=520092 GN=DFR29_111113 PE=4 SV=1
MM1 pKa = 8.0PSTALPRR8 pKa = 11.84LVRR11 pKa = 11.84AWLLISLLSCLPSLAFAQCVSLTTLGSAVTQNFDD45 pKa = 3.8TLSNTAGSTTNNLTITGWFLTEE67 pKa = 4.01SGGGARR73 pKa = 11.84DD74 pKa = 3.55NEE76 pKa = 4.58QYY78 pKa = 11.27GVDD81 pKa = 3.67TGASTTGDD89 pKa = 3.37MYY91 pKa = 11.42SYY93 pKa = 10.86GAAAATDD100 pKa = 3.6RR101 pKa = 11.84ALGQLRR107 pKa = 11.84SGTLIPLFGACFTNNTGSTLASLAVAYY134 pKa = 9.22TGEE137 pKa = 4.12EE138 pKa = 3.98WRR140 pKa = 11.84LGTAARR146 pKa = 11.84TDD148 pKa = 3.64TLAFEE153 pKa = 4.93YY154 pKa = 9.79STTATDD160 pKa = 4.1LVTGAWTGVTALNFVTPDD178 pKa = 3.13TATAGAKK185 pKa = 9.83NGNAAADD192 pKa = 4.1RR193 pKa = 11.84TALASTISGLSIANGASFWIRR214 pKa = 11.84WNDD217 pKa = 3.05TDD219 pKa = 5.77ASGADD224 pKa = 3.97DD225 pKa = 4.92GLAVDD230 pKa = 5.47DD231 pKa = 6.36FSLTPQAPIALPNLSINDD249 pKa = 3.65VILNEE254 pKa = 4.27GNAGTTSFTFTVSLSAPAGPGGVTFDD280 pKa = 3.47IATADD285 pKa = 3.74GTAVAPGDD293 pKa = 4.07YY294 pKa = 8.81VTKK297 pKa = 10.61SLTSQTIPAGSSTYY311 pKa = 11.1SFTVLANGDD320 pKa = 3.79TTPEE324 pKa = 3.95TNEE327 pKa = 3.95TFFVNVTAATGATVTDD343 pKa = 4.34GQGQGTLVNDD353 pKa = 4.13DD354 pKa = 4.11AAPNLTINDD363 pKa = 3.23VSLNEE368 pKa = 4.19GNAGTTTFTFIASLSAPAPAGGVTFDD394 pKa = 3.23IATANGTAIAPGDD407 pKa = 3.8YY408 pKa = 8.67TASSLTGQTIPAGSSTYY425 pKa = 9.85TFSVLVNGDD434 pKa = 3.33ITPEE438 pKa = 3.75SDD440 pKa = 2.91EE441 pKa = 4.49TFVVTITNATNAIVTDD457 pKa = 4.24GQGQGNIVNDD467 pKa = 4.22DD468 pKa = 3.8LSRR471 pKa = 11.84IHH473 pKa = 7.31DD474 pKa = 3.75VQGNGASTPIPGATVTVQGIVIGNFQGAGRR504 pKa = 11.84LQGFFLQEE512 pKa = 3.24ADD514 pKa = 4.01ANADD518 pKa = 3.49ADD520 pKa = 4.08PATSEE525 pKa = 4.85GIFVSCGSCATPVAEE540 pKa = 4.33GQVVRR545 pKa = 11.84ATGVVSEE552 pKa = 4.97SNNLTQISATTAGAVVVTNAGNNLAQVTPTTIDD585 pKa = 3.57LPVVGAIDD593 pKa = 3.88DD594 pKa = 4.41FYY596 pKa = 11.55EE597 pKa = 4.28SRR599 pKa = 11.84EE600 pKa = 4.1SMRR603 pKa = 11.84VTFVDD608 pKa = 3.67TLAVSDD614 pKa = 4.18YY615 pKa = 11.39FEE617 pKa = 4.08MARR620 pKa = 11.84YY621 pKa = 7.99GQLVLMEE628 pKa = 4.71GGRR631 pKa = 11.84PRR633 pKa = 11.84AFTEE637 pKa = 4.15FATPSVAGYY646 pKa = 7.38TAYY649 pKa = 11.05LDD651 pKa = 3.32NLARR655 pKa = 11.84RR656 pKa = 11.84RR657 pKa = 11.84VLLDD661 pKa = 3.92DD662 pKa = 5.46DD663 pKa = 5.4NNAQEE668 pKa = 5.1SFLTQPNGMQSVYY681 pKa = 10.42HH682 pKa = 6.07PRR684 pKa = 11.84ANGGFSFGTQGTDD697 pKa = 3.28FFRR700 pKa = 11.84GGDD703 pKa = 3.8LVNGLTGVLDD713 pKa = 4.54FSDD716 pKa = 4.11PAGAASPSWRR726 pKa = 11.84VRR728 pKa = 11.84PVAATPPTFTVANPRR743 pKa = 11.84PATPPAVGGAIKK755 pKa = 9.7VTGLNLLNYY764 pKa = 7.46FTTIDD769 pKa = 3.57TTASSSAGPCGPGGTLDD786 pKa = 3.76CRR788 pKa = 11.84GADD791 pKa = 3.35SVAEE795 pKa = 4.12LNRR798 pKa = 11.84QRR800 pKa = 11.84EE801 pKa = 4.13RR802 pKa = 11.84ASVVICSLNADD813 pKa = 3.46VAALIEE819 pKa = 4.5LEE821 pKa = 4.19NTTPSATITDD831 pKa = 4.12LLGAINARR839 pKa = 11.84CGGAHH844 pKa = 7.4PYY846 pKa = 10.96AFANTGGTLGTDD858 pKa = 4.58AIRR861 pKa = 11.84VQPIYY866 pKa = 9.62RR867 pKa = 11.84TGIVSPVGSPLSDD880 pKa = 4.2LDD882 pKa = 3.89PVHH885 pKa = 6.41NRR887 pKa = 11.84PPTAQTFDD895 pKa = 3.52VVDD898 pKa = 3.55ATNAAFGQRR907 pKa = 11.84FTVIANHH914 pKa = 6.22LRR916 pKa = 11.84AKK918 pKa = 9.68SCSGATGGDD927 pKa = 3.7ADD929 pKa = 4.18TGDD932 pKa = 3.83GQSCFTAARR941 pKa = 11.84TAQATRR947 pKa = 11.84LLSWISSTVIPAAGDD962 pKa = 3.61PDD964 pKa = 4.2VLLLGDD970 pKa = 4.06FNAYY974 pKa = 9.69AQEE977 pKa = 4.26TPIATIVAGGYY988 pKa = 8.39TDD990 pKa = 5.78LEE992 pKa = 4.64TAFEE996 pKa = 4.62GPAAYY1001 pKa = 10.08SYY1003 pKa = 11.71VFGGALGHH1011 pKa = 7.2LDD1013 pKa = 3.56YY1014 pKa = 11.52AFASASLNSQVIGASAWHH1032 pKa = 6.47INADD1036 pKa = 4.01EE1037 pKa = 4.47VPLLDD1042 pKa = 4.65YY1043 pKa = 10.92NDD1045 pKa = 3.77EE1046 pKa = 4.19VADD1049 pKa = 3.74VGEE1052 pKa = 4.51SAFEE1056 pKa = 3.98EE1057 pKa = 4.66KK1058 pKa = 10.24PDD1060 pKa = 3.67GSALVPPRR1068 pKa = 11.84VVFQPASPYY1077 pKa = 9.82RR1078 pKa = 11.84ASDD1081 pKa = 3.54HH1082 pKa = 7.33DD1083 pKa = 4.15PVLVGLFAVSDD1094 pKa = 3.99LAVTLTDD1101 pKa = 3.45SPDD1104 pKa = 3.43PVVAGNNLTYY1114 pKa = 10.54TITVSNNGPDD1124 pKa = 3.49AATAASWSLSLPPGRR1139 pKa = 11.84SFVSLPAVAGWTCTTPAVGSGGTVSCSNPGFSVGSGVFTLTAAVDD1184 pKa = 3.47PSIAAGTVLTATATVTSASGDD1205 pKa = 3.58PTPGNNSATATTTVGTSADD1224 pKa = 3.49LGVTNSAAPATAISGQAITYY1244 pKa = 7.71TITASNAGPSDD1255 pKa = 3.46AAGVSLSNPLPANTTFASLAAPGGWSCTTPAVGATGTVSCTAASLAVTSAVFTLVVTVDD1314 pKa = 3.47AGTTAANITDD1324 pKa = 3.84TATVSSATSDD1334 pKa = 3.6PNPGNEE1340 pKa = 4.29SATATTPLSAEE1351 pKa = 4.02ADD1353 pKa = 3.55LSVTHH1358 pKa = 6.23VATPLSGPDD1367 pKa = 3.53LAPGEE1372 pKa = 4.46SIQYY1376 pKa = 11.24AMTVTNVGPSSAALVTLSNIMPPQTTLNSLTAPPGWSCTTPALGSNGTISCTRR1429 pKa = 11.84PTLAPGSSTITFVVLLDD1446 pKa = 3.66AGYY1449 pKa = 10.52LGSTVVDD1456 pKa = 3.82TASVTSATTDD1466 pKa = 3.57PAPGNEE1472 pKa = 4.28TQTATTPVLASADD1485 pKa = 3.67LAVSNTASTPTAINGQPITYY1505 pKa = 8.62TVTVTNNGPSNAATVAMNNPIPANTTFTAATTPPGWNCTLPAAGGTGTVACANASLAPGSAVFSITVTVNPGAPVTMIVDD1585 pKa = 4.35TATVSSATGDD1595 pKa = 3.51PATGNEE1601 pKa = 4.37SATATTSTPVSLQSFEE1617 pKa = 4.01VDD1619 pKa = 2.88
MM1 pKa = 8.0PSTALPRR8 pKa = 11.84LVRR11 pKa = 11.84AWLLISLLSCLPSLAFAQCVSLTTLGSAVTQNFDD45 pKa = 3.8TLSNTAGSTTNNLTITGWFLTEE67 pKa = 4.01SGGGARR73 pKa = 11.84DD74 pKa = 3.55NEE76 pKa = 4.58QYY78 pKa = 11.27GVDD81 pKa = 3.67TGASTTGDD89 pKa = 3.37MYY91 pKa = 11.42SYY93 pKa = 10.86GAAAATDD100 pKa = 3.6RR101 pKa = 11.84ALGQLRR107 pKa = 11.84SGTLIPLFGACFTNNTGSTLASLAVAYY134 pKa = 9.22TGEE137 pKa = 4.12EE138 pKa = 3.98WRR140 pKa = 11.84LGTAARR146 pKa = 11.84TDD148 pKa = 3.64TLAFEE153 pKa = 4.93YY154 pKa = 9.79STTATDD160 pKa = 4.1LVTGAWTGVTALNFVTPDD178 pKa = 3.13TATAGAKK185 pKa = 9.83NGNAAADD192 pKa = 4.1RR193 pKa = 11.84TALASTISGLSIANGASFWIRR214 pKa = 11.84WNDD217 pKa = 3.05TDD219 pKa = 5.77ASGADD224 pKa = 3.97DD225 pKa = 4.92GLAVDD230 pKa = 5.47DD231 pKa = 6.36FSLTPQAPIALPNLSINDD249 pKa = 3.65VILNEE254 pKa = 4.27GNAGTTSFTFTVSLSAPAGPGGVTFDD280 pKa = 3.47IATADD285 pKa = 3.74GTAVAPGDD293 pKa = 4.07YY294 pKa = 8.81VTKK297 pKa = 10.61SLTSQTIPAGSSTYY311 pKa = 11.1SFTVLANGDD320 pKa = 3.79TTPEE324 pKa = 3.95TNEE327 pKa = 3.95TFFVNVTAATGATVTDD343 pKa = 4.34GQGQGTLVNDD353 pKa = 4.13DD354 pKa = 4.11AAPNLTINDD363 pKa = 3.23VSLNEE368 pKa = 4.19GNAGTTTFTFIASLSAPAPAGGVTFDD394 pKa = 3.23IATANGTAIAPGDD407 pKa = 3.8YY408 pKa = 8.67TASSLTGQTIPAGSSTYY425 pKa = 9.85TFSVLVNGDD434 pKa = 3.33ITPEE438 pKa = 3.75SDD440 pKa = 2.91EE441 pKa = 4.49TFVVTITNATNAIVTDD457 pKa = 4.24GQGQGNIVNDD467 pKa = 4.22DD468 pKa = 3.8LSRR471 pKa = 11.84IHH473 pKa = 7.31DD474 pKa = 3.75VQGNGASTPIPGATVTVQGIVIGNFQGAGRR504 pKa = 11.84LQGFFLQEE512 pKa = 3.24ADD514 pKa = 4.01ANADD518 pKa = 3.49ADD520 pKa = 4.08PATSEE525 pKa = 4.85GIFVSCGSCATPVAEE540 pKa = 4.33GQVVRR545 pKa = 11.84ATGVVSEE552 pKa = 4.97SNNLTQISATTAGAVVVTNAGNNLAQVTPTTIDD585 pKa = 3.57LPVVGAIDD593 pKa = 3.88DD594 pKa = 4.41FYY596 pKa = 11.55EE597 pKa = 4.28SRR599 pKa = 11.84EE600 pKa = 4.1SMRR603 pKa = 11.84VTFVDD608 pKa = 3.67TLAVSDD614 pKa = 4.18YY615 pKa = 11.39FEE617 pKa = 4.08MARR620 pKa = 11.84YY621 pKa = 7.99GQLVLMEE628 pKa = 4.71GGRR631 pKa = 11.84PRR633 pKa = 11.84AFTEE637 pKa = 4.15FATPSVAGYY646 pKa = 7.38TAYY649 pKa = 11.05LDD651 pKa = 3.32NLARR655 pKa = 11.84RR656 pKa = 11.84RR657 pKa = 11.84VLLDD661 pKa = 3.92DD662 pKa = 5.46DD663 pKa = 5.4NNAQEE668 pKa = 5.1SFLTQPNGMQSVYY681 pKa = 10.42HH682 pKa = 6.07PRR684 pKa = 11.84ANGGFSFGTQGTDD697 pKa = 3.28FFRR700 pKa = 11.84GGDD703 pKa = 3.8LVNGLTGVLDD713 pKa = 4.54FSDD716 pKa = 4.11PAGAASPSWRR726 pKa = 11.84VRR728 pKa = 11.84PVAATPPTFTVANPRR743 pKa = 11.84PATPPAVGGAIKK755 pKa = 9.7VTGLNLLNYY764 pKa = 7.46FTTIDD769 pKa = 3.57TTASSSAGPCGPGGTLDD786 pKa = 3.76CRR788 pKa = 11.84GADD791 pKa = 3.35SVAEE795 pKa = 4.12LNRR798 pKa = 11.84QRR800 pKa = 11.84EE801 pKa = 4.13RR802 pKa = 11.84ASVVICSLNADD813 pKa = 3.46VAALIEE819 pKa = 4.5LEE821 pKa = 4.19NTTPSATITDD831 pKa = 4.12LLGAINARR839 pKa = 11.84CGGAHH844 pKa = 7.4PYY846 pKa = 10.96AFANTGGTLGTDD858 pKa = 4.58AIRR861 pKa = 11.84VQPIYY866 pKa = 9.62RR867 pKa = 11.84TGIVSPVGSPLSDD880 pKa = 4.2LDD882 pKa = 3.89PVHH885 pKa = 6.41NRR887 pKa = 11.84PPTAQTFDD895 pKa = 3.52VVDD898 pKa = 3.55ATNAAFGQRR907 pKa = 11.84FTVIANHH914 pKa = 6.22LRR916 pKa = 11.84AKK918 pKa = 9.68SCSGATGGDD927 pKa = 3.7ADD929 pKa = 4.18TGDD932 pKa = 3.83GQSCFTAARR941 pKa = 11.84TAQATRR947 pKa = 11.84LLSWISSTVIPAAGDD962 pKa = 3.61PDD964 pKa = 4.2VLLLGDD970 pKa = 4.06FNAYY974 pKa = 9.69AQEE977 pKa = 4.26TPIATIVAGGYY988 pKa = 8.39TDD990 pKa = 5.78LEE992 pKa = 4.64TAFEE996 pKa = 4.62GPAAYY1001 pKa = 10.08SYY1003 pKa = 11.71VFGGALGHH1011 pKa = 7.2LDD1013 pKa = 3.56YY1014 pKa = 11.52AFASASLNSQVIGASAWHH1032 pKa = 6.47INADD1036 pKa = 4.01EE1037 pKa = 4.47VPLLDD1042 pKa = 4.65YY1043 pKa = 10.92NDD1045 pKa = 3.77EE1046 pKa = 4.19VADD1049 pKa = 3.74VGEE1052 pKa = 4.51SAFEE1056 pKa = 3.98EE1057 pKa = 4.66KK1058 pKa = 10.24PDD1060 pKa = 3.67GSALVPPRR1068 pKa = 11.84VVFQPASPYY1077 pKa = 9.82RR1078 pKa = 11.84ASDD1081 pKa = 3.54HH1082 pKa = 7.33DD1083 pKa = 4.15PVLVGLFAVSDD1094 pKa = 3.99LAVTLTDD1101 pKa = 3.45SPDD1104 pKa = 3.43PVVAGNNLTYY1114 pKa = 10.54TITVSNNGPDD1124 pKa = 3.49AATAASWSLSLPPGRR1139 pKa = 11.84SFVSLPAVAGWTCTTPAVGSGGTVSCSNPGFSVGSGVFTLTAAVDD1184 pKa = 3.47PSIAAGTVLTATATVTSASGDD1205 pKa = 3.58PTPGNNSATATTTVGTSADD1224 pKa = 3.49LGVTNSAAPATAISGQAITYY1244 pKa = 7.71TITASNAGPSDD1255 pKa = 3.46AAGVSLSNPLPANTTFASLAAPGGWSCTTPAVGATGTVSCTAASLAVTSAVFTLVVTVDD1314 pKa = 3.47AGTTAANITDD1324 pKa = 3.84TATVSSATSDD1334 pKa = 3.6PNPGNEE1340 pKa = 4.29SATATTPLSAEE1351 pKa = 4.02ADD1353 pKa = 3.55LSVTHH1358 pKa = 6.23VATPLSGPDD1367 pKa = 3.53LAPGEE1372 pKa = 4.46SIQYY1376 pKa = 11.24AMTVTNVGPSSAALVTLSNIMPPQTTLNSLTAPPGWSCTTPALGSNGTISCTRR1429 pKa = 11.84PTLAPGSSTITFVVLLDD1446 pKa = 3.66AGYY1449 pKa = 10.52LGSTVVDD1456 pKa = 3.82TASVTSATTDD1466 pKa = 3.57PAPGNEE1472 pKa = 4.28TQTATTPVLASADD1485 pKa = 3.67LAVSNTASTPTAINGQPITYY1505 pKa = 8.62TVTVTNNGPSNAATVAMNNPIPANTTFTAATTPPGWNCTLPAAGGTGTVACANASLAPGSAVFSITVTVNPGAPVTMIVDD1585 pKa = 4.35TATVSSATGDD1595 pKa = 3.51PATGNEE1601 pKa = 4.37SATATTSTPVSLQSFEE1617 pKa = 4.01VDD1619 pKa = 2.88
Molecular weight: 162.65 kDa
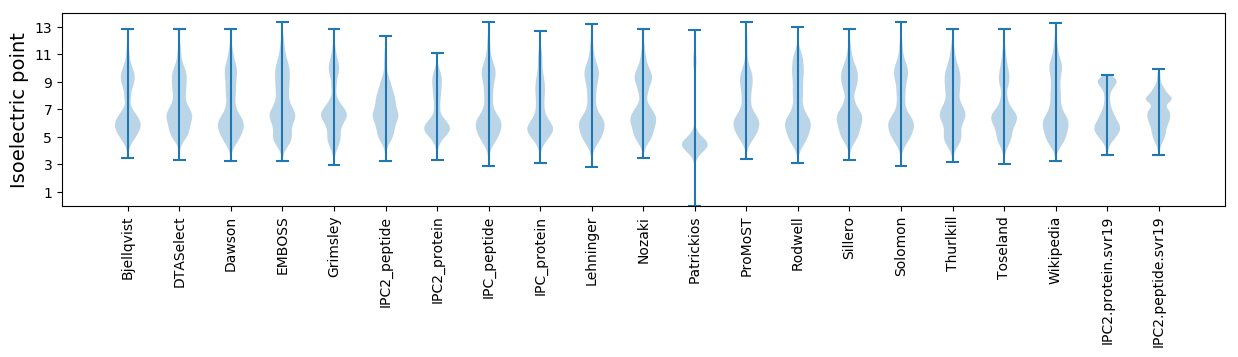
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V3DN13|A0A4V3DN13_9GAMM WD40 repeat protein OS=Tahibacter aquaticus OX=520092 GN=DFR29_103212 PE=4 SV=1
MM1 pKa = 7.58LMLMLGFLLWPLPLPLPLPLPLPLRR26 pKa = 11.84LRR28 pKa = 11.84LRR30 pKa = 11.84LRR32 pKa = 11.84LRR34 pKa = 11.84LLVLVLVLVLVILLWLVILLWLLPLPLLLRR64 pKa = 11.84AGSAPLPGPHH74 pKa = 7.49RR75 pKa = 11.84SRR77 pKa = 4.27
MM1 pKa = 7.58LMLMLGFLLWPLPLPLPLPLPLPLRR26 pKa = 11.84LRR28 pKa = 11.84LRR30 pKa = 11.84LRR32 pKa = 11.84LRR34 pKa = 11.84LLVLVLVLVLVILLWLVILLWLLPLPLLLRR64 pKa = 11.84AGSAPLPGPHH74 pKa = 7.49RR75 pKa = 11.84SRR77 pKa = 4.27
Molecular weight: 8.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2081458 |
27 |
5836 |
367.5 |
39.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.49 ± 0.055 | 0.991 ± 0.011 |
5.67 ± 0.026 | 4.96 ± 0.032 |
3.401 ± 0.019 | 8.35 ± 0.038 |
2.087 ± 0.018 | 4.039 ± 0.021 |
2.519 ± 0.029 | 11.051 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.774 ± 0.016 | 2.818 ± 0.028 |
5.27 ± 0.029 | 4.019 ± 0.023 |
7.521 ± 0.04 | 5.913 ± 0.031 |
5.067 ± 0.044 | 7.118 ± 0.026 |
1.521 ± 0.015 | 2.422 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
