
Amphibacillus xylanus (strain ATCC 51415 / DSM 6626 / JCM 7361 / LMG 17667 / NBRC 15112 / Ep01)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Amphibacillus; Amphibacillus xylanus
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
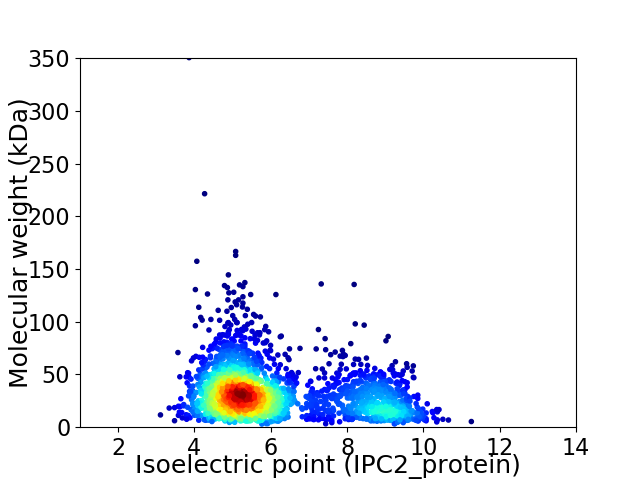
Virtual 2D-PAGE plot for 2384 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
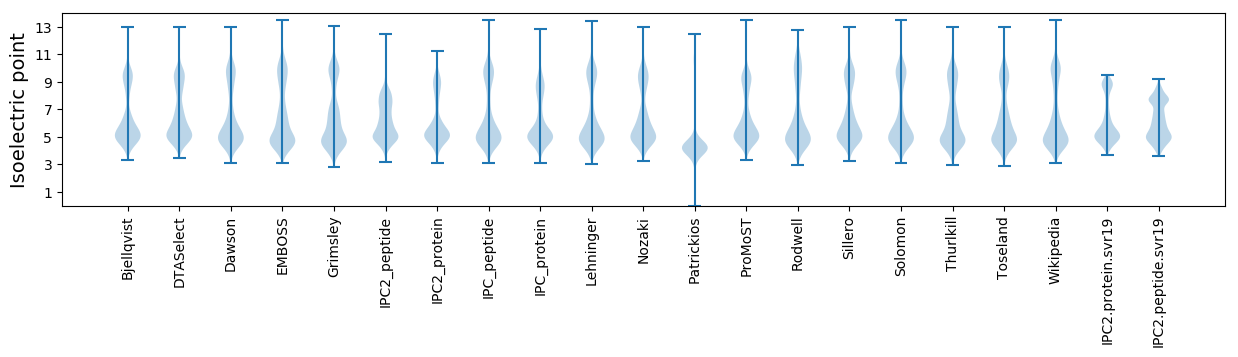
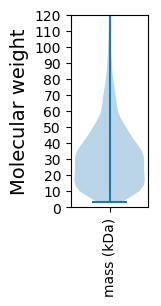
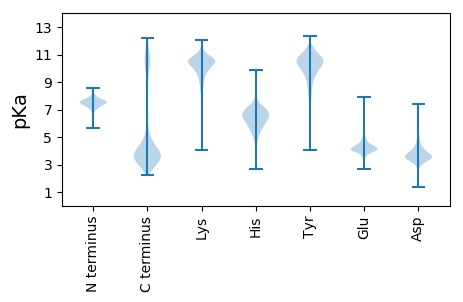
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0J5Y7|K0J5Y7_AMPXN Histidine kinase OS=Amphibacillus xylanus (strain ATCC 51415 / DSM 6626 / JCM 7361 / LMG 17667 / NBRC 15112 / Ep01) OX=698758 GN=AXY_23790 PE=4 SV=1
MM1 pKa = 7.86KK2 pKa = 10.37KK3 pKa = 10.28RR4 pKa = 11.84ILAILLVLIFALVACGDD21 pKa = 4.11KK22 pKa = 10.44DD23 pKa = 3.73TASPNDD29 pKa = 3.47VEE31 pKa = 6.69GDD33 pKa = 3.57DD34 pKa = 3.97TDD36 pKa = 3.87NTEE39 pKa = 3.81EE40 pKa = 4.12TTDD43 pKa = 3.45EE44 pKa = 4.3VEE46 pKa = 4.27EE47 pKa = 4.25EE48 pKa = 4.57EE49 pKa = 4.31PTVDD53 pKa = 4.09YY54 pKa = 10.71EE55 pKa = 4.28QTPEE59 pKa = 3.72MDD61 pKa = 3.35FDD63 pKa = 4.49LGGRR67 pKa = 11.84TLKK70 pKa = 10.71LVSWYY75 pKa = 10.09DD76 pKa = 3.56EE77 pKa = 4.15QVGEE81 pKa = 4.5GSPDD85 pKa = 3.51SIAISEE91 pKa = 4.08NMEE94 pKa = 4.1ALKK97 pKa = 10.45EE98 pKa = 3.91KK99 pKa = 10.82HH100 pKa = 6.33NFDD103 pKa = 3.12IEE105 pKa = 4.65YY106 pKa = 10.59VVVDD110 pKa = 3.17YY111 pKa = 11.59GEE113 pKa = 4.01YY114 pKa = 9.72QEE116 pKa = 5.85RR117 pKa = 11.84VTASLIAGEE126 pKa = 4.09PLGDD130 pKa = 4.21IIRR133 pKa = 11.84LPRR136 pKa = 11.84PWMIPTLTRR145 pKa = 11.84QGLFHH150 pKa = 7.11PVDD153 pKa = 3.47EE154 pKa = 4.47YY155 pKa = 11.83VINEE159 pKa = 3.87NSFVLQYY166 pKa = 10.36TEE168 pKa = 3.75QHH170 pKa = 5.97SEE172 pKa = 3.68FDD174 pKa = 3.19GRR176 pKa = 11.84GYY178 pKa = 10.8GFRR181 pKa = 11.84VGIAGAAGGVFYY193 pKa = 11.15NRR195 pKa = 11.84TLMNEE200 pKa = 4.08LNLDD204 pKa = 3.72PLQAYY209 pKa = 9.1VDD211 pKa = 3.54NGEE214 pKa = 4.4WNWEE218 pKa = 4.03TFKK221 pKa = 11.28AVAEE225 pKa = 4.36SANQDD230 pKa = 3.54TNNDD234 pKa = 3.29GTIDD238 pKa = 3.38TWGLATSSILVQALAANEE256 pKa = 3.74AAIVRR261 pKa = 11.84NGKK264 pKa = 9.52QNLEE268 pKa = 4.11DD269 pKa = 3.93PATIEE274 pKa = 3.75VMEE277 pKa = 5.45FISEE281 pKa = 4.19LGDD284 pKa = 3.87GIARR288 pKa = 11.84PTEE291 pKa = 4.38GGDD294 pKa = 2.9WTEE297 pKa = 3.86PKK299 pKa = 10.58QFFLQGNTLMYY310 pKa = 10.3VGQDD314 pKa = 3.22YY315 pKa = 11.42EE316 pKa = 4.15MADD319 pKa = 4.54FIDD322 pKa = 4.54GLPDD326 pKa = 3.41HH327 pKa = 7.69DD328 pKa = 4.88IGFLPFPMGPSASSYY343 pKa = 6.71QTHH346 pKa = 5.39ITIPNYY352 pKa = 8.74YY353 pKa = 8.8TIPSAVEE360 pKa = 4.02DD361 pKa = 4.09PDD363 pKa = 5.0KK364 pKa = 10.9IVYY367 pKa = 9.19LWEE370 pKa = 3.86KK371 pKa = 9.7MYY373 pKa = 10.55DD374 pKa = 3.05IEE376 pKa = 5.84SIYY379 pKa = 10.51DD380 pKa = 3.53YY381 pKa = 10.98PEE383 pKa = 3.52QANFEE388 pKa = 4.47THH390 pKa = 6.65FNNEE394 pKa = 3.72DD395 pKa = 4.14DD396 pKa = 4.0INNARR401 pKa = 11.84LAVQSFQVIEE411 pKa = 4.18QIDD414 pKa = 4.78YY415 pKa = 8.97YY416 pKa = 10.52PSMPYY421 pKa = 10.36YY422 pKa = 9.8EE423 pKa = 4.76FSGEE427 pKa = 3.96LSDD430 pKa = 4.83GVNISTLIEE439 pKa = 4.22SYY441 pKa = 11.1GPSFQAAIDD450 pKa = 4.16EE451 pKa = 4.64VWGEE455 pKa = 3.9
MM1 pKa = 7.86KK2 pKa = 10.37KK3 pKa = 10.28RR4 pKa = 11.84ILAILLVLIFALVACGDD21 pKa = 4.11KK22 pKa = 10.44DD23 pKa = 3.73TASPNDD29 pKa = 3.47VEE31 pKa = 6.69GDD33 pKa = 3.57DD34 pKa = 3.97TDD36 pKa = 3.87NTEE39 pKa = 3.81EE40 pKa = 4.12TTDD43 pKa = 3.45EE44 pKa = 4.3VEE46 pKa = 4.27EE47 pKa = 4.25EE48 pKa = 4.57EE49 pKa = 4.31PTVDD53 pKa = 4.09YY54 pKa = 10.71EE55 pKa = 4.28QTPEE59 pKa = 3.72MDD61 pKa = 3.35FDD63 pKa = 4.49LGGRR67 pKa = 11.84TLKK70 pKa = 10.71LVSWYY75 pKa = 10.09DD76 pKa = 3.56EE77 pKa = 4.15QVGEE81 pKa = 4.5GSPDD85 pKa = 3.51SIAISEE91 pKa = 4.08NMEE94 pKa = 4.1ALKK97 pKa = 10.45EE98 pKa = 3.91KK99 pKa = 10.82HH100 pKa = 6.33NFDD103 pKa = 3.12IEE105 pKa = 4.65YY106 pKa = 10.59VVVDD110 pKa = 3.17YY111 pKa = 11.59GEE113 pKa = 4.01YY114 pKa = 9.72QEE116 pKa = 5.85RR117 pKa = 11.84VTASLIAGEE126 pKa = 4.09PLGDD130 pKa = 4.21IIRR133 pKa = 11.84LPRR136 pKa = 11.84PWMIPTLTRR145 pKa = 11.84QGLFHH150 pKa = 7.11PVDD153 pKa = 3.47EE154 pKa = 4.47YY155 pKa = 11.83VINEE159 pKa = 3.87NSFVLQYY166 pKa = 10.36TEE168 pKa = 3.75QHH170 pKa = 5.97SEE172 pKa = 3.68FDD174 pKa = 3.19GRR176 pKa = 11.84GYY178 pKa = 10.8GFRR181 pKa = 11.84VGIAGAAGGVFYY193 pKa = 11.15NRR195 pKa = 11.84TLMNEE200 pKa = 4.08LNLDD204 pKa = 3.72PLQAYY209 pKa = 9.1VDD211 pKa = 3.54NGEE214 pKa = 4.4WNWEE218 pKa = 4.03TFKK221 pKa = 11.28AVAEE225 pKa = 4.36SANQDD230 pKa = 3.54TNNDD234 pKa = 3.29GTIDD238 pKa = 3.38TWGLATSSILVQALAANEE256 pKa = 3.74AAIVRR261 pKa = 11.84NGKK264 pKa = 9.52QNLEE268 pKa = 4.11DD269 pKa = 3.93PATIEE274 pKa = 3.75VMEE277 pKa = 5.45FISEE281 pKa = 4.19LGDD284 pKa = 3.87GIARR288 pKa = 11.84PTEE291 pKa = 4.38GGDD294 pKa = 2.9WTEE297 pKa = 3.86PKK299 pKa = 10.58QFFLQGNTLMYY310 pKa = 10.3VGQDD314 pKa = 3.22YY315 pKa = 11.42EE316 pKa = 4.15MADD319 pKa = 4.54FIDD322 pKa = 4.54GLPDD326 pKa = 3.41HH327 pKa = 7.69DD328 pKa = 4.88IGFLPFPMGPSASSYY343 pKa = 6.71QTHH346 pKa = 5.39ITIPNYY352 pKa = 8.74YY353 pKa = 8.8TIPSAVEE360 pKa = 4.02DD361 pKa = 4.09PDD363 pKa = 5.0KK364 pKa = 10.9IVYY367 pKa = 9.19LWEE370 pKa = 3.86KK371 pKa = 9.7MYY373 pKa = 10.55DD374 pKa = 3.05IEE376 pKa = 5.84SIYY379 pKa = 10.51DD380 pKa = 3.53YY381 pKa = 10.98PEE383 pKa = 3.52QANFEE388 pKa = 4.47THH390 pKa = 6.65FNNEE394 pKa = 3.72DD395 pKa = 4.14DD396 pKa = 4.0INNARR401 pKa = 11.84LAVQSFQVIEE411 pKa = 4.18QIDD414 pKa = 4.78YY415 pKa = 8.97YY416 pKa = 10.52PSMPYY421 pKa = 10.36YY422 pKa = 9.8EE423 pKa = 4.76FSGEE427 pKa = 3.96LSDD430 pKa = 4.83GVNISTLIEE439 pKa = 4.22SYY441 pKa = 11.1GPSFQAAIDD450 pKa = 4.16EE451 pKa = 4.64VWGEE455 pKa = 3.9
Molecular weight: 51.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0IUW5|K0IUW5_AMPXN Beta sliding clamp OS=Amphibacillus xylanus (strain ATCC 51415 / DSM 6626 / JCM 7361 / LMG 17667 / NBRC 15112 / Ep01) OX=698758 GN=dnaN PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPKK8 pKa = 8.36NRR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.01KK14 pKa = 7.87VHH16 pKa = 5.71GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.02NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.51KK37 pKa = 10.32GRR39 pKa = 11.84RR40 pKa = 11.84VLSAA44 pKa = 3.76
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPKK8 pKa = 8.36NRR10 pKa = 11.84QRR12 pKa = 11.84KK13 pKa = 8.01KK14 pKa = 7.87VHH16 pKa = 5.71GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTKK25 pKa = 10.02NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 8.51KK37 pKa = 10.32GRR39 pKa = 11.84RR40 pKa = 11.84VLSAA44 pKa = 3.76
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
713264 |
26 |
3128 |
299.2 |
33.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.537 ± 0.049 | 0.568 ± 0.014 |
5.86 ± 0.048 | 7.504 ± 0.061 |
4.359 ± 0.041 | 6.254 ± 0.052 |
2.099 ± 0.024 | 8.524 ± 0.052 |
6.513 ± 0.052 | 9.746 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.474 ± 0.021 | 4.779 ± 0.039 |
3.419 ± 0.035 | 4.401 ± 0.047 |
4.234 ± 0.038 | 5.759 ± 0.03 |
5.498 ± 0.038 | 6.728 ± 0.038 |
0.911 ± 0.02 | 3.833 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
