
Spongiivirga citrea
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Spongiivirga
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
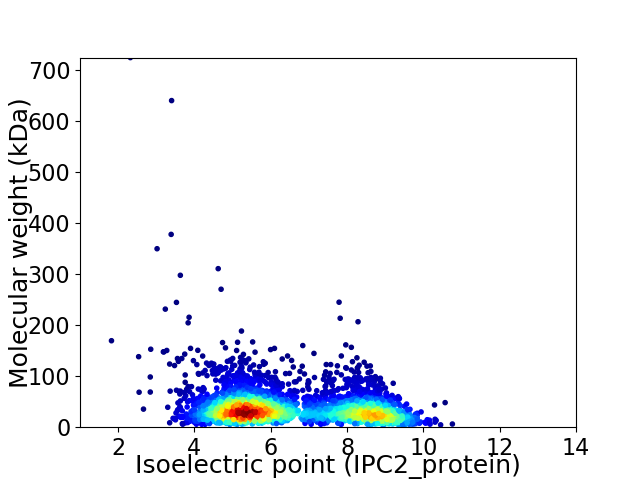
Virtual 2D-PAGE plot for 3588 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M0CLF3|A0A6M0CLF3_9FLAO Cytokinin riboside 5'-monophosphate phosphoribohydrolase OS=Spongiivirga citrea OX=1481457 GN=GWK10_05620 PE=3 SV=1
PP1 pKa = 8.03DD2 pKa = 5.06PFDD5 pKa = 3.84TNVPNTYY12 pKa = 8.88TLTYY16 pKa = 9.93SATGATNVTRR26 pKa = 11.84TVVVNATPVASFTVDD41 pKa = 2.73QSTITEE47 pKa = 3.96GSMVNFDD54 pKa = 4.4ASASTASSAITYY66 pKa = 9.7NWDD69 pKa = 3.38FGDD72 pKa = 4.65GSPIQQTTNPTISHH86 pKa = 6.02QYY88 pKa = 8.83SSNVGSPFTATLSIEE103 pKa = 4.29DD104 pKa = 4.19ANGCTSATNASQEE117 pKa = 4.07ITVNPATSPFIDD129 pKa = 4.74LGALPATVTLCQGAIFTDD147 pKa = 4.02TVSAEE152 pKa = 4.21DD153 pKa = 3.82GNGNIIPVTTTSSPDD168 pKa = 3.41PFDD171 pKa = 3.8TNLPGTYY178 pKa = 9.38TLTYY182 pKa = 9.42SAAGATDD189 pKa = 3.29MTRR192 pKa = 11.84TVVVNTTPVASFNVDD207 pKa = 2.75QLTITEE213 pKa = 4.34GEE215 pKa = 4.43TVNFDD220 pKa = 3.54ASASTASSAITYY232 pKa = 7.45TWNFGDD238 pKa = 4.58GSPIQTTTNPTVDD251 pKa = 3.25HH252 pKa = 6.81TYY254 pKa = 9.03STSAGSPFTTTLTIEE269 pKa = 4.64DD270 pKa = 4.55ADD272 pKa = 3.88GCTNTNTASQEE283 pKa = 3.82IIVNVTGSATVLINSAKK300 pKa = 8.47NASYY304 pKa = 9.09VTEE307 pKa = 4.18NVQITIDD314 pKa = 3.31IDD316 pKa = 3.56EE317 pKa = 4.67TNGGNGPYY325 pKa = 9.04TAGYY329 pKa = 10.08RR330 pKa = 11.84NATGSFDD337 pKa = 3.87LSNGTSINVDD347 pKa = 2.95TDD349 pKa = 3.07ISIGNNITNWTFIGNAAFDD368 pKa = 4.06ASFEE372 pKa = 4.29IYY374 pKa = 10.08VKK376 pKa = 10.84NGAGEE381 pKa = 4.55EE382 pKa = 4.23IGSSTINIAVQTFTINLIPDD402 pKa = 3.99TDD404 pKa = 4.82PINAEE409 pKa = 3.96VGNSVDD415 pKa = 3.82FTLFIGEE422 pKa = 4.23VNGQIGNQYY431 pKa = 10.66NITINTDD438 pKa = 2.5QGTDD442 pKa = 3.27GTINAGGDD450 pKa = 3.54SSTGTLTINNQSPGSPISISYY471 pKa = 10.3IGTTATNHH479 pKa = 6.55AISVSVVPVSFNGLSRR495 pKa = 11.84NEE497 pKa = 3.93NRR499 pKa = 11.84NINYY503 pKa = 7.69TSTAFPFTFSASTVTVNPFLFFNNEE528 pKa = 2.75IDD530 pKa = 3.33INLNITPAGPVGGGQHH546 pKa = 5.4EE547 pKa = 4.5VRR549 pKa = 11.84FTGDD553 pKa = 3.08LAGGQILDD561 pKa = 3.63QSNNTVSLNTWTSVNNSNLTVWKK584 pKa = 9.64FRR586 pKa = 11.84AGTSIGIADD595 pKa = 4.48LNFEE599 pKa = 4.59ARR601 pKa = 11.84NASGGTVTGNPQNINIKK618 pKa = 10.27IIDD621 pKa = 3.84FSITVNPTTTITEE634 pKa = 4.21TAGTQVTNLSVRR646 pKa = 11.84IDD648 pKa = 3.54PTDD651 pKa = 3.06TSFISAYY658 pKa = 10.37DD659 pKa = 3.37LTIASDD665 pKa = 3.49QADD668 pKa = 4.34GIFNSGDD675 pKa = 3.22THH677 pKa = 6.17ITTTVNGGASFNFNYY692 pKa = 10.23TGTSVNLHH700 pKa = 6.04NLSISATPVGQTEE713 pKa = 4.98PIRR716 pKa = 11.84TSAFRR721 pKa = 11.84IDD723 pKa = 4.01YY724 pKa = 10.23VAGNQPPIAQNDD736 pKa = 3.83SFIIQQNSSQNLLLVYY752 pKa = 10.32ADD754 pKa = 3.8NGSGIDD760 pKa = 4.12EE761 pKa = 4.96DD762 pKa = 4.71PEE764 pKa = 5.4NDD766 pKa = 4.22DD767 pKa = 5.77FIINQIPSNPSKK779 pKa = 11.02GITAIQGDD787 pKa = 4.84AINYY791 pKa = 8.33VPTNGEE797 pKa = 3.91FGNDD801 pKa = 2.7TFTYY805 pKa = 10.26NLEE808 pKa = 4.74DD809 pKa = 3.31INGNVSNDD817 pKa = 2.87ATVSVFINRR826 pKa = 11.84PPTINPQSVTRR837 pKa = 11.84LISDD841 pKa = 3.8RR842 pKa = 11.84SGTLLISINSLTNDD856 pKa = 3.73PDD858 pKa = 3.35GHH860 pKa = 5.67TVLLNAVGTPSNNGGTAIIGTGQQTIVFSGNGGTWATSTFTYY902 pKa = 10.07QVSDD906 pKa = 3.83GFGGTATGTVIVEE919 pKa = 4.37FDD921 pKa = 4.06PGTGPDD927 pKa = 3.93PCDD930 pKa = 3.27NCPPGTSCTNCDD942 pKa = 2.86ICISPMQACPQAPPKK957 pKa = 10.29EE958 pKa = 4.01GRR960 pKa = 11.84NPIKK964 pKa = 10.55VEE966 pKa = 3.87KK967 pKa = 9.9VNKK970 pKa = 9.84GKK972 pKa = 10.74AKK974 pKa = 10.11IEE976 pKa = 4.11KK977 pKa = 9.55QQ978 pKa = 3.17
PP1 pKa = 8.03DD2 pKa = 5.06PFDD5 pKa = 3.84TNVPNTYY12 pKa = 8.88TLTYY16 pKa = 9.93SATGATNVTRR26 pKa = 11.84TVVVNATPVASFTVDD41 pKa = 2.73QSTITEE47 pKa = 3.96GSMVNFDD54 pKa = 4.4ASASTASSAITYY66 pKa = 9.7NWDD69 pKa = 3.38FGDD72 pKa = 4.65GSPIQQTTNPTISHH86 pKa = 6.02QYY88 pKa = 8.83SSNVGSPFTATLSIEE103 pKa = 4.29DD104 pKa = 4.19ANGCTSATNASQEE117 pKa = 4.07ITVNPATSPFIDD129 pKa = 4.74LGALPATVTLCQGAIFTDD147 pKa = 4.02TVSAEE152 pKa = 4.21DD153 pKa = 3.82GNGNIIPVTTTSSPDD168 pKa = 3.41PFDD171 pKa = 3.8TNLPGTYY178 pKa = 9.38TLTYY182 pKa = 9.42SAAGATDD189 pKa = 3.29MTRR192 pKa = 11.84TVVVNTTPVASFNVDD207 pKa = 2.75QLTITEE213 pKa = 4.34GEE215 pKa = 4.43TVNFDD220 pKa = 3.54ASASTASSAITYY232 pKa = 7.45TWNFGDD238 pKa = 4.58GSPIQTTTNPTVDD251 pKa = 3.25HH252 pKa = 6.81TYY254 pKa = 9.03STSAGSPFTTTLTIEE269 pKa = 4.64DD270 pKa = 4.55ADD272 pKa = 3.88GCTNTNTASQEE283 pKa = 3.82IIVNVTGSATVLINSAKK300 pKa = 8.47NASYY304 pKa = 9.09VTEE307 pKa = 4.18NVQITIDD314 pKa = 3.31IDD316 pKa = 3.56EE317 pKa = 4.67TNGGNGPYY325 pKa = 9.04TAGYY329 pKa = 10.08RR330 pKa = 11.84NATGSFDD337 pKa = 3.87LSNGTSINVDD347 pKa = 2.95TDD349 pKa = 3.07ISIGNNITNWTFIGNAAFDD368 pKa = 4.06ASFEE372 pKa = 4.29IYY374 pKa = 10.08VKK376 pKa = 10.84NGAGEE381 pKa = 4.55EE382 pKa = 4.23IGSSTINIAVQTFTINLIPDD402 pKa = 3.99TDD404 pKa = 4.82PINAEE409 pKa = 3.96VGNSVDD415 pKa = 3.82FTLFIGEE422 pKa = 4.23VNGQIGNQYY431 pKa = 10.66NITINTDD438 pKa = 2.5QGTDD442 pKa = 3.27GTINAGGDD450 pKa = 3.54SSTGTLTINNQSPGSPISISYY471 pKa = 10.3IGTTATNHH479 pKa = 6.55AISVSVVPVSFNGLSRR495 pKa = 11.84NEE497 pKa = 3.93NRR499 pKa = 11.84NINYY503 pKa = 7.69TSTAFPFTFSASTVTVNPFLFFNNEE528 pKa = 2.75IDD530 pKa = 3.33INLNITPAGPVGGGQHH546 pKa = 5.4EE547 pKa = 4.5VRR549 pKa = 11.84FTGDD553 pKa = 3.08LAGGQILDD561 pKa = 3.63QSNNTVSLNTWTSVNNSNLTVWKK584 pKa = 9.64FRR586 pKa = 11.84AGTSIGIADD595 pKa = 4.48LNFEE599 pKa = 4.59ARR601 pKa = 11.84NASGGTVTGNPQNINIKK618 pKa = 10.27IIDD621 pKa = 3.84FSITVNPTTTITEE634 pKa = 4.21TAGTQVTNLSVRR646 pKa = 11.84IDD648 pKa = 3.54PTDD651 pKa = 3.06TSFISAYY658 pKa = 10.37DD659 pKa = 3.37LTIASDD665 pKa = 3.49QADD668 pKa = 4.34GIFNSGDD675 pKa = 3.22THH677 pKa = 6.17ITTTVNGGASFNFNYY692 pKa = 10.23TGTSVNLHH700 pKa = 6.04NLSISATPVGQTEE713 pKa = 4.98PIRR716 pKa = 11.84TSAFRR721 pKa = 11.84IDD723 pKa = 4.01YY724 pKa = 10.23VAGNQPPIAQNDD736 pKa = 3.83SFIIQQNSSQNLLLVYY752 pKa = 10.32ADD754 pKa = 3.8NGSGIDD760 pKa = 4.12EE761 pKa = 4.96DD762 pKa = 4.71PEE764 pKa = 5.4NDD766 pKa = 4.22DD767 pKa = 5.77FIINQIPSNPSKK779 pKa = 11.02GITAIQGDD787 pKa = 4.84AINYY791 pKa = 8.33VPTNGEE797 pKa = 3.91FGNDD801 pKa = 2.7TFTYY805 pKa = 10.26NLEE808 pKa = 4.74DD809 pKa = 3.31INGNVSNDD817 pKa = 2.87ATVSVFINRR826 pKa = 11.84PPTINPQSVTRR837 pKa = 11.84LISDD841 pKa = 3.8RR842 pKa = 11.84SGTLLISINSLTNDD856 pKa = 3.73PDD858 pKa = 3.35GHH860 pKa = 5.67TVLLNAVGTPSNNGGTAIIGTGQQTIVFSGNGGTWATSTFTYY902 pKa = 10.07QVSDD906 pKa = 3.83GFGGTATGTVIVEE919 pKa = 4.37FDD921 pKa = 4.06PGTGPDD927 pKa = 3.93PCDD930 pKa = 3.27NCPPGTSCTNCDD942 pKa = 2.86ICISPMQACPQAPPKK957 pKa = 10.29EE958 pKa = 4.01GRR960 pKa = 11.84NPIKK964 pKa = 10.55VEE966 pKa = 3.87KK967 pKa = 9.9VNKK970 pKa = 9.84GKK972 pKa = 10.74AKK974 pKa = 10.11IEE976 pKa = 4.11KK977 pKa = 9.55QQ978 pKa = 3.17
Molecular weight: 102.22 kDa
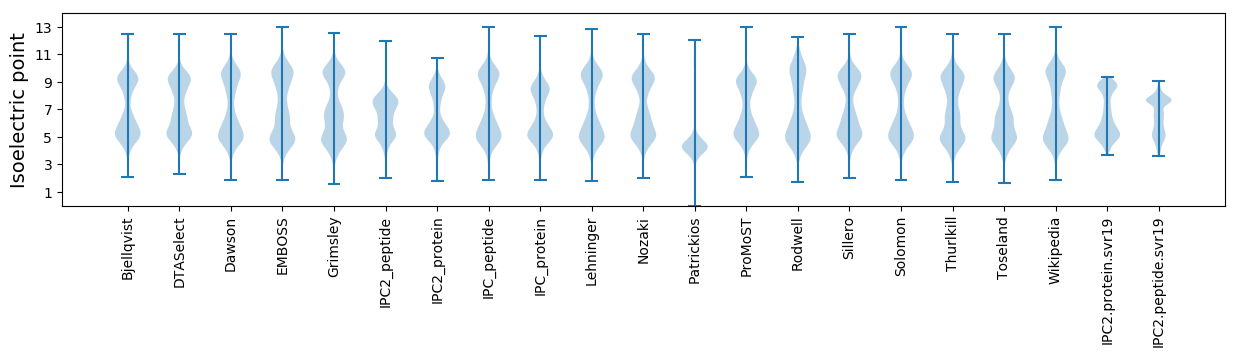
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M0CSK8|A0A6M0CSK8_9FLAO Type IX secretion system membrane protein PorP/SprF (Fragment) OS=Spongiivirga citrea OX=1481457 GN=GWK10_16855 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.17RR21 pKa = 11.84MASVSGRR28 pKa = 11.84KK29 pKa = 9.06VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.52KK41 pKa = 10.3ISVSSDD47 pKa = 2.85PRR49 pKa = 11.84HH50 pKa = 6.1KK51 pKa = 10.6KK52 pKa = 9.83
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.17RR21 pKa = 11.84MASVSGRR28 pKa = 11.84KK29 pKa = 9.06VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.92GRR39 pKa = 11.84KK40 pKa = 8.52KK41 pKa = 10.3ISVSSDD47 pKa = 2.85PRR49 pKa = 11.84HH50 pKa = 6.1KK51 pKa = 10.6KK52 pKa = 9.83
Molecular weight: 6.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1259079 |
34 |
7025 |
350.9 |
39.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.575 ± 0.041 | 0.727 ± 0.015 |
5.995 ± 0.057 | 6.357 ± 0.042 |
5.198 ± 0.035 | 6.638 ± 0.047 |
1.693 ± 0.025 | 7.718 ± 0.036 |
7.346 ± 0.086 | 9.12 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.028 | 6.086 ± 0.042 |
3.438 ± 0.033 | 3.464 ± 0.022 |
3.575 ± 0.036 | 6.538 ± 0.032 |
6.038 ± 0.102 | 6.383 ± 0.043 |
1.087 ± 0.016 | 3.887 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
